Abstract
Diamond-Blackfan anemia (DBA) is associated with developmental defects and profound anemia. Mutations in genes encoding a ribosomal protein of the small (eg, RPS19) or large (eg, RPL11) ribosomal subunit are found in more than half of these patients. The mutations cause ribosomal haploinsufficiency, which reduces overall translation efficiency of cellular mRNAs. We reduced the expression of Rps19 or Rpl11 in mouse erythroblasts and investigated mRNA polyribosome association, which revealed deregulated translation initiation of specific transcripts. Among these were Bag1, encoding a Hsp70 cochaperone, and Csde1, encoding an RNA-binding protein, and both were expressed at increased levels in erythroblasts. Their translation initiation is cap independent and starts from an internal ribosomal entry site, which appeared sensitive to knockdown of Rps19 or Rpl11. Mouse embryos lacking Bag1 die at embryonic day 13.5, with reduced erythroid colony forming cells in the fetal liver, and low Bag1 expression impairs erythroid differentiation in vitro. Reduced expression of Csde1 impairs the proliferation and differentiation of erythroid blasts. Protein but not mRNA expression of BAG1 and CSDE1 was reduced in erythroblasts cultured from DBA patients. Our data suggest that impaired internal ribosomal entry site–mediated translation of mRNAs expressed at increased levels in erythroblasts contributes to the erythroid phenotype of DBA.
Introduction
Diamond-Blackfan anemia (DBA) presents as normochromic, macrocytic anemia with reduced erythroid precursors in the BM.1 Approximately half of DBA patients have skeletal abnormalities such as thumb malformations and growth retardation.2 DBA is mostly diagnosed in infants less than 1 year of age, but in recent years, nonclassic cases of DBA are being diagnosed in adult patients.1
DBA is associated with mutations in genes encoding ribosomal proteins in 55% of patients.3 The most prominently mutated gene (in 25% of patients) is RPS19,4 but mutations in RPS7, RPS10, RPS17, RPS24, and RPS26 in the small ribosomal subunit and in RPL5, RPL11, and RPL35A in the large ribosomal subunit have also been found.3 The mutations cause haploinsufficiency of ribosomal proteins and lead to loss of ribosome function; this reduces general translation, as observed in lymphocytes derived from DBA patients.5 Knockdown of RPS19 in hematopoietic progenitors either from human BM or cord blood decreases the colony-forming capacity of erythroid progenitors, whereas it affects the colony-forming capacity of myeloid progenitors to a far lesser extent.6 Knockdown of Rps19 in mouse fetal liver–derived erythroblasts impairs their proliferation, but the differentiation of cells that survive the knockdown is not affected.7
Because ribosome synthesis consumes up to 25% of a cell's energy, a disbalance in the synthesis of ribosomal proteins activates p53 and inhibits cell proliferation.8 Free Rpl11 and Rpl5 bind and inhibit Mdm2, which reduces p53 ubiquitination and leads to its stabilization. Erythroid cells may be more sensitive to p53 activation than other hematopoietic lineages,9 which is also exemplified by the requirement to inactivate p53 in Friend virus–induced erythroleukemia.10 In a zebrafish model of DBA, however, inactivation of Tp53 rescued the morphological abnormalities caused by morpholinos against RPS19, but did not impair erythropoiesis.11,12 Therefore, it is possible that ribosomal insufficiency causes a specific defect in erythroblasts.
The expansion and maturation of erythroblasts depends on erythropoietin (Epo) and SCF,13,14 and requires activation of the PI3K pathway.15 This results in phosphorylation and activation of mammalian target of rapamycin (mTOR), which phosphorylates 4E-binding protein (4EBP), subsequently releases the cap-binding eukaryotic initiation factor 4E (eIF4E), and allows for association of the translation scanning complex.16 We have shown previously that expansion of erythroblasts depends on the SCF-induced availability of eIF4E, which is required for the translation of specific transcripts with a complex RNA structure such as Igbp1 and Use1.17 The scanning complex includes the small ribosomal subunit, and the 60S subunit joins at the AUG start codon.16 We hypothesized that the reduced availability of ribosomal subunits in DBA, similar to the availability of eIF4E, may affect translation initiation of specific transcripts important for erythroid development.
To investigate the role of translation in a DBA model independently of p53 activation, we reduced Rps19 and Rpl11 expression in the p53-deficient mouse erythroblast line I/11 and examined mRNA polyribosome association. Loss of Rps19 or Rpl11 suppressed the translation of a specific set of transcripts, including Bag1 and Csde1. Their mRNA expression was high in erythroblasts, and their 5′-untranslated region (5′UTR) contains an internal ribosomal entry site (IRES). Protein levels of CSDE1 and BAG1 were also low in erythroblasts cultured from the peripheral blood of DBA patients, whereas RNA expression was not affected. Mouse embryos deficient in the Hsp70/Hsc70 cochaperone Bag1 die at embryonic day 13.5 (E13.5) with a lack of definitive erythrocytes and a marked reduction of fetal liver erythroid progenitors. Reduced expression of the RNA-binding protein Csde1 inhibits both proliferation and maturation of erythroblasts. In summary, these data provide the first evidence for an erythroid-specific mechanism that contributes to the anemia seen in DBA.
Methods
Cell culture
I/11 erythroblasts derived from p53-deficient mouse fetal livers or primary fetal liver-derived erythroblasts were expanded in StemPro-34 medium (Invitrogen) supplemented with 0.5 U/mL of Epo (a gift from Janssen-Cilag), 100 ng/mL of SCF (supernatant of CHO [Chinese hamster ovary] producer cells), and 10−6 M dexamethasone (Sigma-Aldrich).15 For differentiation, medium was supplemented with 5 U/mL of Epo and 0.5 mg/mL of iron-loaded transferrin (Scipac). Cell number and size were determined with an electronic cell counter (CASY-1; Innovatis). Hek293T cells were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FCS (PAA Laboratories). After approval from the Ethics Committee of Faculty Hospital, Palacky University, and informed consent, 30 mL of patient blood was harvested in heparin tubes and erythroblasts were cultured as described previously.18
Hemoglobin content and cell morphology
Cell morphology was assessed by cytospins stained with histological dyes and neutral benzidine17 using a 40× magnification microscope (Leica). Images were processed in Adobe Photoshop CS3 Version 10.0.
Plasmids, transfections, and bicistronic assays
The pβgal/CAT and pβgal-LamB1-CAT were kindly provided by W. Mikulits (Medical University, Vienna, Austria). 5′UTRs of Bag1 and Csde1 were amplified from mouse cDNA using the Fidelity PCR kit (Roche). Amplicons were subcloned using the TA cloning kit (Invitrogen), sequence verified, and cloned in pβgal/CAT by NheI and XhoI sites. The CAT activity was determined 48 hours after transfection using the CAT ELISA assay (Roche). Transfections of siRNA pools against RPS19, RPL11, and scrambled control (Dharmacon) were done with INTERFERin (Polyplus Transfection) according to the manufacturer's protocol.
Lentivirus production, titration, and transductions
Cells (293HekT) were transfected with pLKO.1-puro lentiviral construct containing shRNA sequences (MISSION TRC-Mm 1.0 shRNA library; Sigma-Aldrich; supplemental Table 4, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), pMD2.G, and pSPAX.2 packaging plasmids (gift of T. van Dijk, Erasmus MC, Rotterdam, The Netherlands) using LT-1 transfection reagent (Mirus Bio). Medium was concentrated by ultracentrifugation at 60 000g for 2 hours at 4°C. Lentivirus titer was assessed by colony assays in 293HekT cells. Erythroblasts were transduced with a multiplicity of infection of 3-5 and addition of 8 μg/mL of Polybrene (Sigma-Aldrich). Cells were selected with 1 μg/ml puromycin 24 hours after transduction.
Sucrose gradients and polyribosomal profiles
I/11 cells (35 × 106) were incubated with 0.1 mg/mL of cycloheximide for 10 minutes at 37°C, washed with cold PBS containing 0.1 mg/mL of cycloheximide, and lysed as described previously.17 Polysomal RNA was separated on 10.5-mL 7%-46% linear sucrose gradients containing 10mM Tris-HCl, 12mM MgCl2, and 140mM NaCl, and centrifuged at 190 000g for 3 hours. Measurement of absorbance at 254 nm, visualization, and fractionation were done with the Econo System (Bio-Rad). RNA structure was predicted using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).19
RNA isolation, cDNA synthesis, and qRT-PCR
In vitro–synthesized luciferase RNA was added to each fraction of the sucrose gradient to control for RNA-isolation efficiency from the distinct fractions. RNA was isolated from gradient fractions as described previously,17 and further purified with LiCl.20 Total RNA was isolated using the RNeasy Mini or Micro (for BM cells) Kit (QIAGEN). RNA from BM was amplified with the MessageAmp II aRNA kit (Ambion). After amplification, 1.6 μg of RNA was used for cDNA preparation with random hexamers and the SuperScript II Reverse Transcriptase kit (Invitrogen). Quantitative RT-PCR (qRT-PCR) was performed as described previously.17 Primers are listed in supplemental Table 4. For mouse erythroblasts, β-tubulin was used as a control; for human erythroblasts, B2M selected from a set of 32 endogenous controls was used (Applied Biosystems).
SDS-PAGE and Western blotting
Cell lysates, SDS-PAGE, and Western blotting were performed as described previously.17 Abs used were Rps19 (ab40833; Abcam), Rpl11 (sc-25931; Santa Cruz Biotechnology), Bag1 (sc-939; Santa Cruz Biotechnology), Csde1 (13319-1-AP; ProteinTech Group), Rpl8 (ab55952; Abcam), β-actin (sc-1616; Santa Cruz Biotechnology), and Erk1/2 (sc-94; Santa Cruz Biotechnology). Fluorescently labeled secondary Abs were used and blots were scanned on an Odyssey infrared scanner (LI-COR).
Mice, flow cytometry, and colony assays
The generation of Bag1-knockout mice was described previously.21 All mice were housed in the central animal facilities of the University of Würzburg. The animal care and ethic committee approved all procedures and experiments. The fetal liver cells from E12.5 embryos were resuspended in PBS containing 2% BSA and 0.5 μg/mL of 7-amino-actinomycin D and stained with Abs against Ter119 and CD71 (BD Biosciences). For BFU-E colony assay, fetal liver cells were cultured in MethoCult 3231 (StemCell Technologies) supplemented with murine SCF (50 ng/mL), murine IL-3 (100 ng/mL, purified from supernatant of WEHI cells), and human Epo (4 U/mL). For GM-CSF-–induced colony formation, cells were cultured in MethoCult 3231 supplemented with murine GM-CSF (50 ng/mL). Cytokines were purchased from PeproTech unless stated otherwise. Colonies were visualized with an inverted bright-field microscope and were assigned scores on day 10 (BFU-E) or day 5 (CFU-GM). All colony assays were done in triplicate with seeding at 2 × 104 cells per plate.
RNA-protein pull-down
MEL cells (100 × 106) were used for the RNA-protein pull-down, as described previously.22 For pull-downs, streptavidin-conjugated Dynabeads M-270 (Invitrogen) were used. The NT2 washing buffer was supplied with 0.5% DOCH. RNA was isolated using TRIzol (Invitrogen) and amplified with the MessageAmp II amplification kit (Ambion).
DNA microarrays and data processing
Complementary RNA was generated from pooled subpolyribosomal RNA and polyribosomal RNA fractions as described previously,17 and hybridized to Affymetrix MOE 430A2.0 mouse expression arrays according to the manufacturer's protocols. Each sample had 2 paired hybridizations for the polyribosomal and subpolyribosomal fractions. The raw data were processed using Affymetrix Microarray Suite 5 (MAS5). Probe sets that had a polyribosomal intensity < 100 were excluded to prevent systematic variation between experimental conditions. Subsequently, the intensity values were normalized per probe set for each sample within the associated class to quantify the mRNA transcript association with polyribosomes. These are computed by taking the polyribosomal intensity value per probe set of each sample with respect to the “polysomal plus subpolysomal” intensity value within the sample. This resulted in arbitrary ratios that range between 0 and 1. Data files were uploaded to the Gene Expression Omnibus as accession number GSE22903.
Bioinformatical analyses were performed using Matlab R2009b (MathWorks). To determine transcripts with differential ratios of polysome recruitment, a multivariate analysis using ANOVA was performed using the translational ratios. The multiple testing was controlled by using a Benjamini-Hochberg correction.
Statistical analysis
Results are depicted as means ± SEM unless stated otherwise. Statistical analysis was performed with the Student t test using Prism Version 4 software (GraphPad).
Results
Loss of Rps19 and Rpl11 impairs expansion and differentiation of erythroblasts
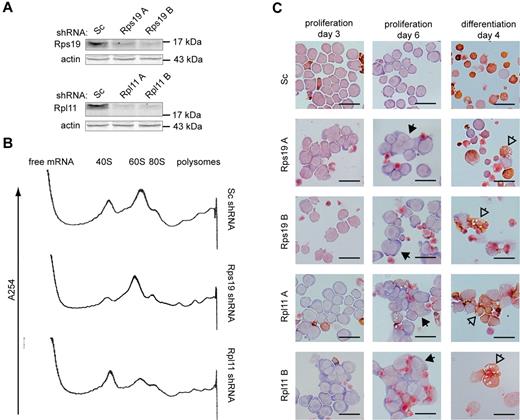
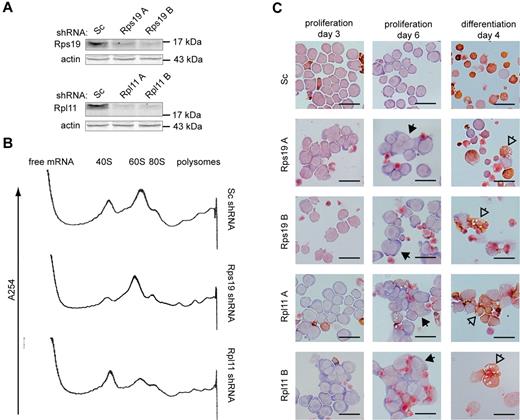
To investigate how haploinsufficiency of ribosomal proteins affects mRNA translation in erythroblasts, the expression of Rps19 and Rpl11 in mouse p53-deficient I/11 erythroblasts was reduced using 2 distinct shRNA sequences for each ribosomal protein (Figure 1A). Single ribosomal subunits, monosomes, and polysomes were separated on a 7%-46% sucrose gradient (supplemental Figure 1A). At day 3, knockdown of Rps19 reduced the 40S subunit content and loss of Rpl11 reduced the 60S subunits (Figure 1B). At this stage, cell numbers and cell morphology between cultures transduced with control shRNA or shRNAs directed against Rps19 or Rpl11 were still comparable (Figure 1C and supplemental Figure 1B). Thereafter, reduced expression of Rps19 and Rpl11 inhibited proliferation of I/11 erythroblasts and resulted in aberrant cell morphology characterized by the occurrence of binucleated or polynucleated cells, depending on which shRNA was used (Figure 1C and supplemental Figure 1B-C). Rps19- and Rpl11-deficient erythroblasts failed to undergo differentiation divisions, accumulated less hemoglobin, and displayed aberrant vacuolized morphology (Figure 1C). In summary, the knockdown of Rps19 and Rpl11 resulted in functional reduction of corresponding ribosomal subunits, followed by phenotypical changes in erythroblasts during proliferation and differentiation.
Phenotype of Rps19- and Rpl11-deficient erythroblasts. (A) Rps19 and Rpl11 protein levels in I/11 erythroblasts 3 days after transduction with lentiviral vectors expressing scrambled (Sc) shRNA or 2 distinct shRNAs (A-B) specific for Rps19 or Rpl11, respectively. (B) RNA profiles measured by absorbance at 254 nm in a sucrose gradient loaded with cytoplasmic extract of shRNA-treated erythroblasts 3 days after transduction. The positions of the 40S and 60S ribosomal subunits, the 80S monosome, and polysomes are indicated (for peak identification, see supplemental Figure 1). (C) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days (left panel) and 6 days (middle panel) in proliferation conditions or switched to differentiation conditions 3 days after transduction and differentiated for 4 days (right panel). Cells were stained for hemoglobin (brown) and histological dyes. Full arrowheads point to multinucleated cells; empty arrowheads point to aberrantly differentiated cells. Scale bars represent 25 μm.
Phenotype of Rps19- and Rpl11-deficient erythroblasts. (A) Rps19 and Rpl11 protein levels in I/11 erythroblasts 3 days after transduction with lentiviral vectors expressing scrambled (Sc) shRNA or 2 distinct shRNAs (A-B) specific for Rps19 or Rpl11, respectively. (B) RNA profiles measured by absorbance at 254 nm in a sucrose gradient loaded with cytoplasmic extract of shRNA-treated erythroblasts 3 days after transduction. The positions of the 40S and 60S ribosomal subunits, the 80S monosome, and polysomes are indicated (for peak identification, see supplemental Figure 1). (C) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days (left panel) and 6 days (middle panel) in proliferation conditions or switched to differentiation conditions 3 days after transduction and differentiated for 4 days (right panel). Cells were stained for hemoglobin (brown) and histological dyes. Full arrowheads point to multinucleated cells; empty arrowheads point to aberrantly differentiated cells. Scale bars represent 25 μm.
Reduced Rps19 expression impairs translation of a specific set of transcripts
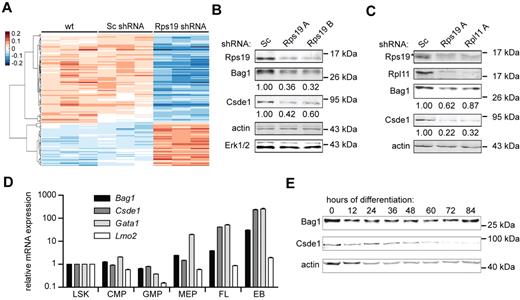
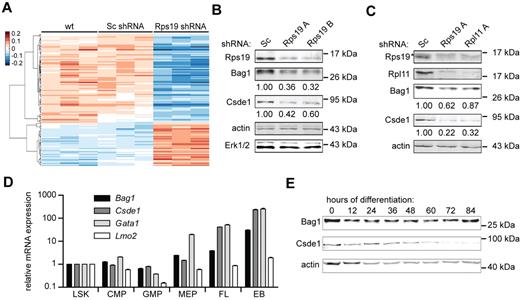
To identify transcripts for which translation may be specifically affected by the loss of Rps19, oligonucleotide arrays were hybridized with cRNA derived from polyribosomal and subpolyribosomal RNA of the nontransduced wild-type cells and cells transduced with lentiviral vectors expressing control shRNA (Sc) or shRNA directed to Rps19 or Rpl11. Arrays hybridized to polysomal or subpolysomal RNA were normalized separately and arbitrary translation ratios were computed for each paired sample to quantify mRNA ribosome loading as follows: (polysomal signal) / (polysomal + subpolysomal signal; see “DNA microarrays and data processing”). Multivariate analysis (false discovery rate < 0.05) identified 134 probe sets, corresponding to 130 genes that were differentially recruited to polysomes after Rps19 knockdown compared with control samples (Figure 2A and supplemental Table 1). Most of these transcripts were regulated similarly on Rps19 or Rpl11 knockdown (supplemental Figure 2A). Transcripts that were selectively lost from polyribosomes encoded, among others, mitochondrial proteins, translation initiation factors, and mRNA-processing factors. We measured the actual percentage of polysome recruitment by qRT-PCR, for which transcripts were selected that had increased expression in erythroid progenitors compared with other hematopoietic lineages23 or for which a predicted RNA structure in the 5′UTR may explain regulated polysome association.19,24 After Rps19 knockdown, polysome recruitment of Bcl2-associated athanogene 1 (Bag1) mRNA decreased from 70% to 40%, cold shock domain containing E1 (Csde1) from 50% to 20%, fracture callus 1 (Fxc1) from 75% to 20%, and Siva1 from 70% to 30%. A control transcript (Hdhd2) retained 65% polyribosome association. Loss of Rpl11 reduced polyribosome recruitment of Bag1, Csde1, Fxc1, and Siva1 to a lesser extent than loss of Rps19, whereas the presence or absence of growth factor signaling (Epo and SCF) did not alter polyribosome recruitment (supplemental Figure 2B). Protein levels of Bag1 (the predominant p36 or the Bag-1S isoform) and Csde1 were correlated with polyribosome recruitment in p53-deficient I/11 cells transduced with control or Rps19 shRNA (Figure 2B; Fxc1 and Siva could not be detected on Western blot with available Abs). These effects were not specific for Rps19, because reduced Rpl11 expression also decreased Bag1 and Csde1 protein expression (supplemental Figure 2C). Knockdown of Rps19 and Rpl11 expression in primary mouse fetal liver erythroblasts confirmed reduced Bag1 and Csde1 protein levels in p53-proficient cells, whereas RNA expression remained constant (Figure 2C and supplemental Figure 2D). Therefore, the loss of Rps19 and Rpl11 negatively affects translation of Bag1 and Csde1 transcripts independently of p53.
Polysome association of mRNAs in erythroblasts deficient for Rps19 and Rpl11. (A) Polysomal- and subpolysomal-derived cRNA generated from parental I/11 erythroblasts (wt), and cells transduced with Sc shRNA or Rps19 shRNA was hybridized to expression arrays. After normalization, arbitrary ratios of polyribosomal recruitment were calculated (see “DNA microarrays and data processing”). Transcripts that are differentially expressed between control cells (wild-type and Sc shRNA) and Rps19-deficient cells were identified by ANOVA (false discovery rate < 0.05) and hierarchically clustered. Three columns for each class represent independent biologic replicates. (B) Protein levels of Bag1 and Csde1 in I/11 erythroblasts 3 days after transduction with Sc and Rps19 shRNA. Actin and Erk1/2 served as loading controls. Quantified expression, corrected for actin, is indicated below the blots. (C) Protein levels of Rps19, Rpl11, Bag1, and Csde1 in primary erythroblasts obtained from E12.5 fetal liver cells 3 days after transduction with Sc, Rps19A, and Rpl11A shRNA. Actin served as a control. Numbers indicate blot quantifications corrected to actin. Asterisk indicates a nonspecific band in the Rps19 panel. (D) qRT-PCR on RNA obtained from sorted mouse BM fractions, day E13.5 mouse fetal liver (FL), and 5 days in vitro erythroid culture of the fetal liver cells (EB, erythroblasts). LSK indicates Lin−Sca1+c-Kit+; CMP, common myeloid progenitor (Lin−c-Kit+CD34+CD16/CD32low); GMP, granulocyte-monocytic progenitor (Lin−c-Kit+CD34+CD16/CD32high); MEP, megakaryocytic-erythroid progenitor (Lin−c-Kit+CD34−CD16/CD32low). Expression is shown relative to LSK levels on a logarithmic scale. Error bars represent ± SEM (n = 4). (E) Mouse erythroblasts (I/11) were induced to differentiate. Every 12 hours, protein samples were prepared to be analyzed for expression of Bag1 and Csde1. Actin served as a loading control. The positions of size markers are indicated.
Polysome association of mRNAs in erythroblasts deficient for Rps19 and Rpl11. (A) Polysomal- and subpolysomal-derived cRNA generated from parental I/11 erythroblasts (wt), and cells transduced with Sc shRNA or Rps19 shRNA was hybridized to expression arrays. After normalization, arbitrary ratios of polyribosomal recruitment were calculated (see “DNA microarrays and data processing”). Transcripts that are differentially expressed between control cells (wild-type and Sc shRNA) and Rps19-deficient cells were identified by ANOVA (false discovery rate < 0.05) and hierarchically clustered. Three columns for each class represent independent biologic replicates. (B) Protein levels of Bag1 and Csde1 in I/11 erythroblasts 3 days after transduction with Sc and Rps19 shRNA. Actin and Erk1/2 served as loading controls. Quantified expression, corrected for actin, is indicated below the blots. (C) Protein levels of Rps19, Rpl11, Bag1, and Csde1 in primary erythroblasts obtained from E12.5 fetal liver cells 3 days after transduction with Sc, Rps19A, and Rpl11A shRNA. Actin served as a control. Numbers indicate blot quantifications corrected to actin. Asterisk indicates a nonspecific band in the Rps19 panel. (D) qRT-PCR on RNA obtained from sorted mouse BM fractions, day E13.5 mouse fetal liver (FL), and 5 days in vitro erythroid culture of the fetal liver cells (EB, erythroblasts). LSK indicates Lin−Sca1+c-Kit+; CMP, common myeloid progenitor (Lin−c-Kit+CD34+CD16/CD32low); GMP, granulocyte-monocytic progenitor (Lin−c-Kit+CD34+CD16/CD32high); MEP, megakaryocytic-erythroid progenitor (Lin−c-Kit+CD34−CD16/CD32low). Expression is shown relative to LSK levels on a logarithmic scale. Error bars represent ± SEM (n = 4). (E) Mouse erythroblasts (I/11) were induced to differentiate. Every 12 hours, protein samples were prepared to be analyzed for expression of Bag1 and Csde1. Actin served as a loading control. The positions of size markers are indicated.
Because DBA is mainly manifested in the erythroid lineage, the expression of Bag1 and Csde1 was compared in various hematopoietic lineages. The expression of Bag1 and Csde1 was low in stem cells and myeloid progenitors, but increased in megakaryocytic-erythroid progenitors and E13.5 fetal liver cells (90% Ter119+ cells) and reached expression levels in cultured erythroblasts that were 30- and 250-fold higher, respectively, compared with levels detected in stem cells (Figure 2D). The increase in Csde1 expression was similar to the increase of the erythroid-specific transcription factor Gata1, which is abundantly expressed in erythroid progenitors. During terminal erythroid differentiation, the expression of Bag1 remained constant throughout differentiation. Csde1 expression was down-regulated 60 hours after induction of differentiation, which corresponds to the start of enucleation15 (Figure 2E). The increased expression of Bag1 and Csde1 throughout the late stages of erythropoiesis suggests an erythroid-specific function.
Expression of BAG1 and CSDE1 is reduced in DBA patient–derived erythroblasts
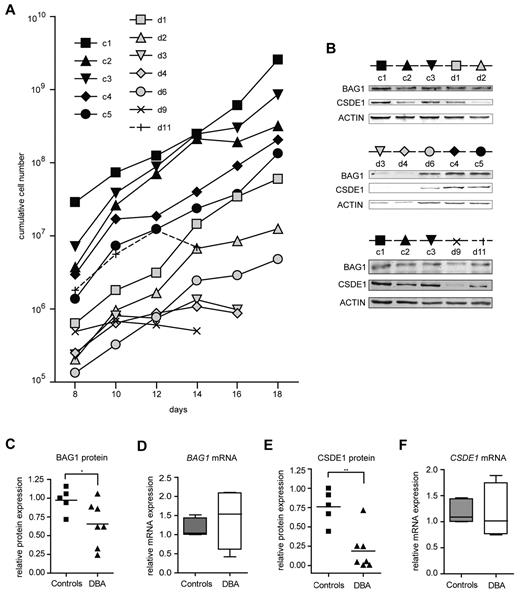
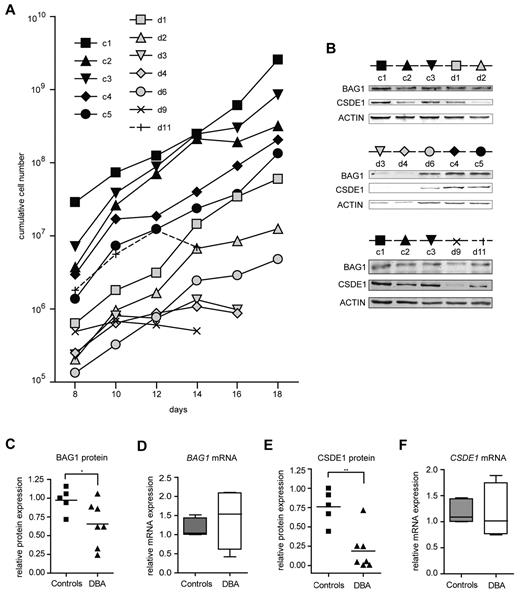
To examine whether the identified genes are target genes in DBA independent of the underlying mutation, we assessed the expression of BAG1 and CSDE1 in erythroblasts cultured from mononuclear cells isolated from 30 mL of peripheral blood from DBA patients with various mutations and from age- and sex-matched controls (supplemental Table 2). In 7 of 11 patient samples, we could expand erythroblasts, albeit with a total yield that was reduced compared with healthy controls (Figure 3A). The low quantity of patient-derived erythroblasts precluded the isolation of polysomes from sucrose gradients. Therefore, we harvested protein and RNA samples from the cultures to examine protein expression on Western blots, which we then related to the mRNA expression measured by qRT-PCR (for quantitative data, see supplemental Table 3). BAG1 protein levels were reduced in DBA samples compared with controls (Figure 3B-C), whereas BAG1 mRNA levels remained constant or were even increased (Figure 3D). Expression of CSDE1 protein was on average 3-fold lower in DBA erythroblasts than in control erythroblasts (Figure 3B,E), whereas CSDE1 mRNA levels were similar between DBA and controls (Figure 3F). To exclude the possibility that the differences in CSDE1 levels resulted from the differentiation status of the cultures, we stained the cells at the day of protein and RNA harvest (supplemental Figure 3). Together the results indicated that defective translation of specific targets occurs in DBA patient–derived erythroblasts as well, similar to the mouse in vitro cultures. Interestingly, reduced proliferation rates of DBA erythroblast cultures seemed to be correlated with low BAG1 and CSDE1 protein levels (compare Figure 3A-B).
BAG1 and CSDE1 protein expression is decreased in DBA erythroblasts. (A) Proliferation of erythroblasts derived from peripheral mononuclear blood of DBA patients and healthy controls. Cumulative cell numbers were calculated; days after in vitro culture initiation are indicated. Black symbols represent healthy controls (c1-c5); gray or open symbols represent DBA patients (d1-d11). Erythroblasts of 5 healthy controls and 7 DBA patients were harvested on day 16 of in vitro culture for protein lysates and RNA isolation; only samples d9 and d11 were harvested on day 14. (B) Protein expression of BAG1 and CSDE1 was analyzed by Western blot using actin as a loading control. The symbols above the Western blot correspond to symbols used in panel A. Patient characteristics are given in supplemental Table 2. BAG1 (C) and CSDE1 (E) protein expression was quantified and corrected to actin expression. BAG1 (D) and CSDE1 (F) transcript levels were quantified by qRT-PCR (except patient d9, n = 4 per sample). Data are also given in supplemental Table 3, lines indicate mean in panels C through F; boxes in panels D and F indicate SEM. Error bars indicate maximum and minimum values. *P = .059; **P = .0023.
BAG1 and CSDE1 protein expression is decreased in DBA erythroblasts. (A) Proliferation of erythroblasts derived from peripheral mononuclear blood of DBA patients and healthy controls. Cumulative cell numbers were calculated; days after in vitro culture initiation are indicated. Black symbols represent healthy controls (c1-c5); gray or open symbols represent DBA patients (d1-d11). Erythroblasts of 5 healthy controls and 7 DBA patients were harvested on day 16 of in vitro culture for protein lysates and RNA isolation; only samples d9 and d11 were harvested on day 14. (B) Protein expression of BAG1 and CSDE1 was analyzed by Western blot using actin as a loading control. The symbols above the Western blot correspond to symbols used in panel A. Patient characteristics are given in supplemental Table 2. BAG1 (C) and CSDE1 (E) protein expression was quantified and corrected to actin expression. BAG1 (D) and CSDE1 (F) transcript levels were quantified by qRT-PCR (except patient d9, n = 4 per sample). Data are also given in supplemental Table 3, lines indicate mean in panels C through F; boxes in panels D and F indicate SEM. Error bars indicate maximum and minimum values. *P = .059; **P = .0023.
Bag1 deficiency impairs erythroid differentiation
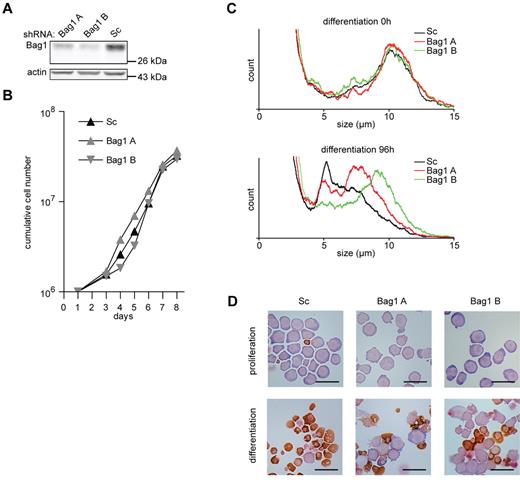
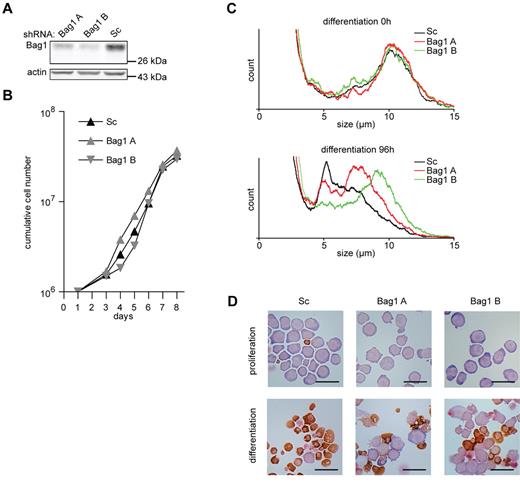
To study the function of Bag1 in erythroid differentiation, different lentiviral shRNA expressing constructs were used to reduce Bag1 expression in I/11 erythroblasts to 50% and 25% of normal levels (Figure 4A). Reduced Bag1 expression did not affect the expansion of erythroblast cultures (Figure 4B). However, upon induction of differentiation, which was characterized by size reduction and hemoglobin accumulation, a considerable subset of Bag1-depleted erythroblasts retained their size and morphology (Figure 4C-D). Inhibition of differentiation was inversely correlated with the expression of Bag1 at the beginning of differentiation. Not all cells were retained in an immature state, and cells able to enter the differentiation program accumulated hemoglobin at a level comparable to control cells (Figure 4D).
Bag1 is required for I/11 erythroid differentiation. (A) Protein levels of Bag1 in I/11 erythroblasts 3 days after transduction with Sc shRNA lentivirus and 2 distinct shRNA lentiviruses complementary to Bag1. (B) Cumulative cell numbers of I/11 erythroblast cultures after transduction with control shRNA (▴) and shRNA against Bag1 (A  and B
and B  ) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.
) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.
Bag1 is required for I/11 erythroid differentiation. (A) Protein levels of Bag1 in I/11 erythroblasts 3 days after transduction with Sc shRNA lentivirus and 2 distinct shRNA lentiviruses complementary to Bag1. (B) Cumulative cell numbers of I/11 erythroblast cultures after transduction with control shRNA (▴) and shRNA against Bag1 (A  and B
and B  ) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.
) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.
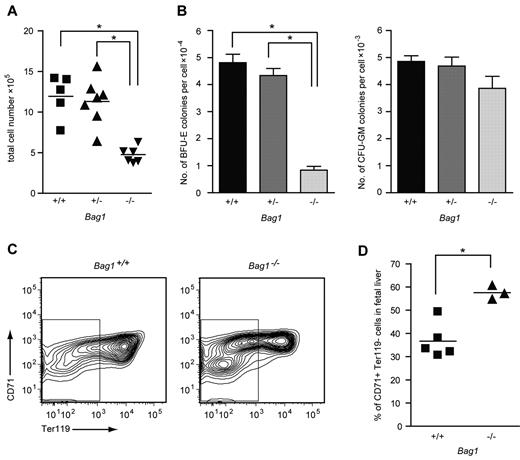
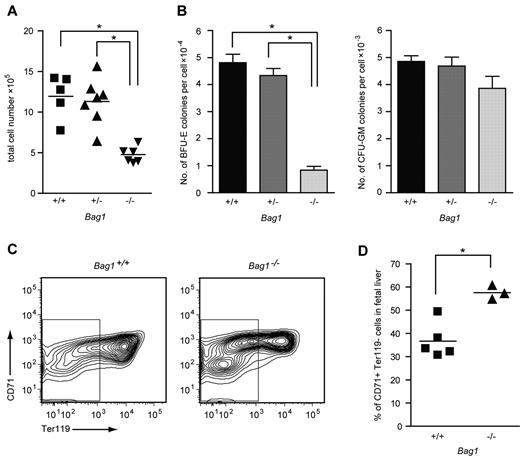
Bag1−/− mouse embryos died at day E13.5 with severe defects in brain development and small, anemic livers.21 To investigate the hematopoietic defect in Bag1−/− livers, we examined fetal liver cells from E12.5 Bag1−/− embryos and Bag1+/+ and Bag1+/− littermates. Bag1−/− fetal livers contained half the number of cells compared with Bag1+/+ and Bag1+/− fetal livers (Figure 5A). In colony-forming assays, 104Bag1−/− fetal liver cells yielded 4 times fewer BFU-E colonies compared with Bag1+/+ and Bag1+/− fetal liver cells, whereas the number of CFU-GM colonies was not significantly different between the genotypes (Figure 5B). To analyze maturation of Bag1−/− fetal liver cells, we measured expression of the erythroid markers CD71 and Ter119 on the fetal liver cells. The Bag1−/− fetal livers contained a TER119−, CD71low population of immature cells that was barely present in wild-type fetal livers. Within the CD71high erythroid population, fewer Bag1−/− fetal liver cells coexpressed TER119, indicating delayed differentiation (Figure 5C-D). These data suggest that complete loss of Bag1 expression strongly impairs erythropoiesis. Fewer BFU-E colonies were present in the fetal livers at day E12.5, and the erythroid progenitors that were generated were delayed in their maturation to TER119+ cells. When Bag1 expression is reduced in established erythroblasts, they are less prone to enter terminal differentiation, but part of the culture matures to hemoglobinated cells. We conclude that Bag1 is required for erythroblasts to enter a terminal differentiation program.
Bag1 deficiency inhibits maturation of erythroblasts in vivo. (A) Total number of nucleated cells in E12.5 fetal livers from Bag1+/+, Bag1+/−, and Bag1−/− embryos. *P < .001. (B) Number of colonies in BFU-E and CFU-GM assays represented as colonies per cell from E12.5 fetal livers, as in panel A. *P < .001. (C-D) FACS analysis of the 7AAD− cells from fetal livers, as in panel A. The gated area represents the Ter119− cells, which are quantified as the percentage of the total in the graph in panel D. *P < .005.
Bag1 deficiency inhibits maturation of erythroblasts in vivo. (A) Total number of nucleated cells in E12.5 fetal livers from Bag1+/+, Bag1+/−, and Bag1−/− embryos. *P < .001. (B) Number of colonies in BFU-E and CFU-GM assays represented as colonies per cell from E12.5 fetal livers, as in panel A. *P < .001. (C-D) FACS analysis of the 7AAD− cells from fetal livers, as in panel A. The gated area represents the Ter119− cells, which are quantified as the percentage of the total in the graph in panel D. *P < .005.
Loss of Csde1, an IRES-binding protein, inhibits erythroid proliferation and differentiation
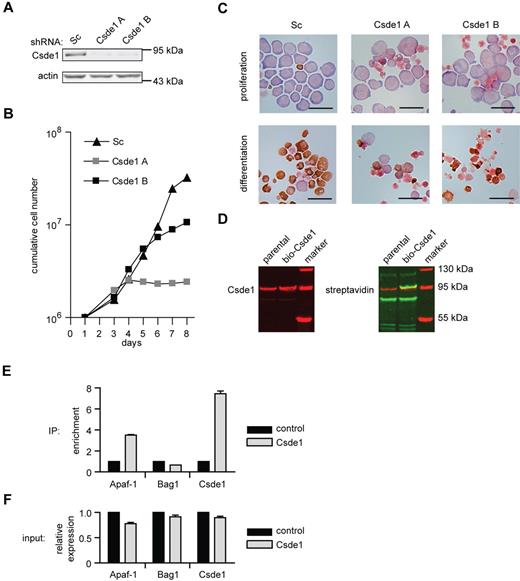
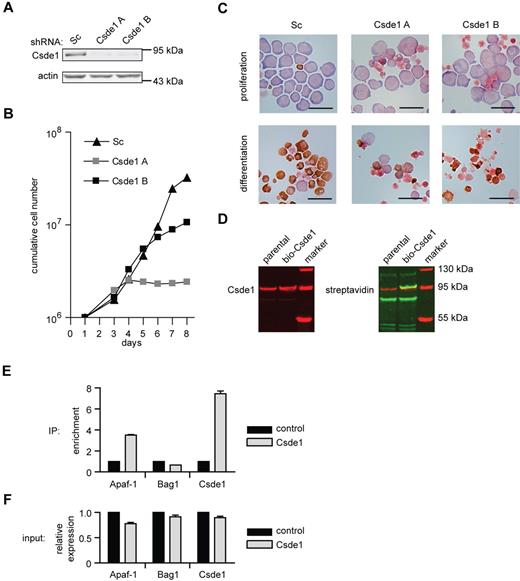
Csde1 contains 5 copies of the RNA-binding cold-shock domain and is highly homologous with Drosophila upstream of N-ras (unr), which is involved in mRNA translation.25 We suppressed Csde1 expression in I/11 erythroblasts using lentiviral vectors expressing 2 distinct shRNAs. Expression was reduced to 10% (Csde1 shRNA-A) or 20% (Csde1 shRNA-B) of normal protein levels (Figure 6A), which impaired both erythroid proliferation and differentiation (Figure 6B-C). The cells with reduced Csde1 expression became mainly pyknotic and failed to mature to enucleated erythrocytes (Figure 6C).
Csde1 is required for proliferation and differentiation of I/11 erythroblasts. (A) Protein levels of Csde1 in I/11 erythroblasts 3 days after transduction with Sc shRNA lentivirus and 2 distinct shRNA lentiviruses complementary to Csde1. (B) Cumulative cell numbers of I/11 erythroblast cultures after transduction with control shRNA (▴) and shRNA against Csde1 (A ▩ and B ■). Numbers are calculated from 3 independent experiments. (C) I/11 erythroblasts were transduced with lentiviral shRNA as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm. (D) Western blots containing lysates from parental MEL-birA cells and cells expressing bio-tagged Csde1 were stained for Csde1 (red). The tagged band runs above the endogenous protein (left image) and was stained with fluorophore-conjugated streptavidin, indicating biotinylation (yellow-green band, right image). (E) The enrichment of Apaf-1, Bag1, and Csde1 in RNA isolated from Csde1 pull-downs with streptavidin beads from MEL cells expressing biotagged-Csde11 compared with pull-downs from parental MEL cells (set at 1; n = 4). (F) Real-time PCR on total RNA used as input in RNA-IP (n = 4).
Csde1 is required for proliferation and differentiation of I/11 erythroblasts. (A) Protein levels of Csde1 in I/11 erythroblasts 3 days after transduction with Sc shRNA lentivirus and 2 distinct shRNA lentiviruses complementary to Csde1. (B) Cumulative cell numbers of I/11 erythroblast cultures after transduction with control shRNA (▴) and shRNA against Csde1 (A ▩ and B ■). Numbers are calculated from 3 independent experiments. (C) I/11 erythroblasts were transduced with lentiviral shRNA as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm. (D) Western blots containing lysates from parental MEL-birA cells and cells expressing bio-tagged Csde1 were stained for Csde1 (red). The tagged band runs above the endogenous protein (left image) and was stained with fluorophore-conjugated streptavidin, indicating biotinylation (yellow-green band, right image). (E) The enrichment of Apaf-1, Bag1, and Csde1 in RNA isolated from Csde1 pull-downs with streptavidin beads from MEL cells expressing biotagged-Csde11 compared with pull-downs from parental MEL cells (set at 1; n = 4). (F) Real-time PCR on total RNA used as input in RNA-IP (n = 4).
Translation of both Csde1 and Bag1 was shown to start from an IRES.26,27 Interestingly, Csde1 protein binds to an IRES sequence in the 5′UTR of specific transcripts such as Apaf-1 and Cdk11A or Rhinovirus RNA.25 It also binds to the IRES in the 5′UTR of its own transcript, and thereby it regulates its own translation in an autoregulatory loop.28 We investigated whether Csde1 protein binds its own IRES and the IRES of Bag1 in erythroblasts. Mouse Csde1 was fused to the recognition site of the prokaryotic biotin ligase BirA at its N-terminus, and the biotagged Csde1 protein was expressed in murine erythroleukemia (MEL) cells that stably expressed the biotin ligase.29 Biotagged Csde1 was efficiently biotinylated (Figure 6D). We then used streptavidin-coated beads to pull down Csde1 and the associated RNA. RNA was converted to cDNA and tested for the presence of Csde1 nucleotide sequences (primers in 3′UTR) with real-time PCR. Csde1 was 8-fold increased in pull-downs from MEL cells coexpressing BirA and biotagged Csde1 compared with MEL cells expressing BirA only. The known target, Apaf-1, was also enriched in Csde1 immunoprecipitations, but Bag1 was not enriched (Figure 6E). Conversely, input mRNA levels before pull-down were comparable between control and Csde1 MEL cells (Figure 6F), suggesting that Csde1 is not involved in the IRES-mediated translation of Bag1 in erythroid cells.
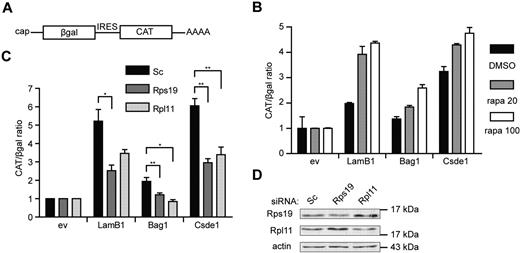
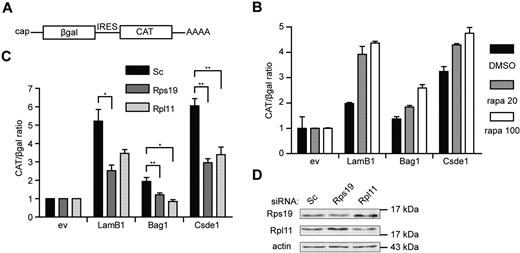
Finally, we investigated whether knockdown of Rps19 and Rpl11 can affect IRES-mediated translation initiation in general. For this, we used a pβgal/CAT bicistronic reporter plasmid,30 in which we inserted the 5′UTR of Bag1 and Csde1 between the open reading frames of β-galactosidase (βgal) and chloramphenicol acetyl transferase (CAT; Figure 7A). The LamB1 5′UTR was used as a positive control for eukaryotic IRES activity.31 Cap-independent translation from the IRES of LamB1, Bag1, and Csde1 was confirmed by an increased CAT/βgal ratio in transfected Hek239T cells after treatment with rapamycin (Figure 7B). The CAT/βgal ratio activity of all constructs was decreased in cells transfected with RPS19 or RPL11 siRNA compared with cells transfected with control siRNA, indicating that the loss of RPS19 and RPL11 decreased IRES-driven translation (Figure 7C). Knockdown of RPS19 and RPL11 in Hek293T cells was confirmed by Western blot (Figure 7D). These data indicate that IRES-mediated translation is sensitive to deficiencies in Rps19 or Rpl11 to a larger extent than cap-dependent translation. Because the IRES activity of the positive control was also affected, we suspect that these phenomena may be a general feature of translation driven by a cellular IRES.
IRES activity is inhibited under ribosomal protein deficiency. (A) Schematic representation of the transcript produced by the bicistronic reporter plasmid with cap-dependent βgal and IRES-dependent CAT expression. (B) Hek293T cells were pretreated with 20 and 100nM rapamycin (gray and white bars, respectively) or with DMSO (black bars) for 12 hours. Cells were transfected with a βgal-CAT bicistronic reporter plasmid lacking IRES sequence (ev) or harboring the 5′UTR of LamB1, Bag1, and Csde1 between the βgal and CAT open reading frame. Cells were harvested 48 hours after transfection, and βgal and CAT were measured using ELISA assays. All βgal-CAT ratios were normalized to the ratio in lysate containing the bicistronic construct without IRES, which was set to 1 in each condition. Error bars indicate SEM. (n = 4). (C) Hek293T cells were treated with Scrambled (Sc), Rps19, and Rpl11 siRNA and transfected after 12 hours with the same bicistronic plasmids. IRES activity is represented by the CAT/βgal ratio. Data shown are means ± SEM (n = 4). *P < .05; **P < .01. (D) Western blot showing knockdown of RPS19 and RPL11 by siRNA in Hek293T cells 3 days after transfection.
IRES activity is inhibited under ribosomal protein deficiency. (A) Schematic representation of the transcript produced by the bicistronic reporter plasmid with cap-dependent βgal and IRES-dependent CAT expression. (B) Hek293T cells were pretreated with 20 and 100nM rapamycin (gray and white bars, respectively) or with DMSO (black bars) for 12 hours. Cells were transfected with a βgal-CAT bicistronic reporter plasmid lacking IRES sequence (ev) or harboring the 5′UTR of LamB1, Bag1, and Csde1 between the βgal and CAT open reading frame. Cells were harvested 48 hours after transfection, and βgal and CAT were measured using ELISA assays. All βgal-CAT ratios were normalized to the ratio in lysate containing the bicistronic construct without IRES, which was set to 1 in each condition. Error bars indicate SEM. (n = 4). (C) Hek293T cells were treated with Scrambled (Sc), Rps19, and Rpl11 siRNA and transfected after 12 hours with the same bicistronic plasmids. IRES activity is represented by the CAT/βgal ratio. Data shown are means ± SEM (n = 4). *P < .05; **P < .01. (D) Western blot showing knockdown of RPS19 and RPL11 by siRNA in Hek293T cells 3 days after transfection.
Discussion
Since the discovery that mutations in ribosomal proteins cause DBA, it has been puzzling why such a general cellular defect predominantly causes profound anemia. Our previous findings that selective mRNA translation is crucial in the expansion of the erythroid compartment prompted us to investigate whether loss of ribosomal proteins impairs the translation of specific transcripts with an essential role in the proliferation or differentiation of erythroblasts. We identified transcripts that were selectively lost from polysomes after knockdown of Rps19 or Rpl11 compared with control erythroblasts. Among these are Bag1 and Csde1, transcripts that are strongly up-regulated in erythroid cells compared with other hematopoietic cells and that appear to be essential for erythropoiesis. Both Bag1 and Csde1 are translated from an IRES and, interestingly, Csde1 is an RNA-binding factor that controls IRES-mediated translation. However, Bag1 mRNA is not a target of Csde1 regulation. Instead, reduced expression of Rps19 or Rpl11 suppressed general IRES-mediated translation more profoundly than cap-dependent translation.
Ribosomal proteins are functional constituents of the ribosome, used, for example, to shape the mRNA exit channel.32 Ribosomal proteins also function in the maturation of ribosomal subunits, and knockdown of Rps19 was shown to affect the maturation of 21S to 18SE pre-rRNA.33 The scattered location of DBA-associated ribosomal proteins on the 40S ribosomal subunit supports the suggested role in ribogenesis, rather than a specific functional role in translation.34 Cap-dependent and IRES-mediated translation compete for translation factors, a process in which cap-dependent translation is favored under optimal conditions. Inhibition of cap-dependent translation by cell starvation, viral infection, or apoptosis induction increases IRES-dependent translation.35 A shortage of ribosomes, as occurs in DBA, may affect the less-competitive mechanism of IRES-mediated translation much more than cap-dependent translation. A more quantitative approach should reveal whether all IRES-mediated translation is affected or whether some IRES-containing transcripts are affected more than others.
We did not just identify transcripts that specifically failed translation after knockdown of Rps19 and Rpl11, some transcripts seemed to be preferentially translated. Among these were transcripts encoding ribosomal proteins (Rps8, Rps25, Rpl8, and Rpl36a) and other transcripts with a 5′-terminal oligopyrimidine tract sequence in their 5′UTR, such as the proteasome subunits Psmb1 and Psmb4. Moreover, we found preferential translation of transcripts that depend on SCF-induced PI3K activation (eg, Use1 and Nap1l1).17 The translation of these transcripts is under the control of the mTOR pathway, and mTOR activity also enhances the translation of 5′-terminal oligopyrimidine tract mRNAs.36 After Rps19 and Rpl11 knockdown, we harvested polysomal RNA from cells cultured in the presence of SCF. Under these conditions, the translation of transcripts with a structured 5′UTR is actively supported. The most likely explanation for the preferential translation of these transcripts is that their translation is still sustained by SCF-dependent mechanisms when the overall translation is reduced because of loss of ribosomes.
Mouse embryos lacking Bag1 die at E13.5, suffering from a major defect in brain development and lack of erythropoiesis. Embryos lacking Epo or the EpoR also die at day 13.5 from a lack of erythrocytes, which indicates that the mouse embryo becomes dependent on definitive erythropoiesis at day E13.5.13 The fetal liver of Bag1−/− embryos was devoid of late erythroid cells, suggesting that the embryos died from impaired erythropoiesis. Surprisingly, we were able to expand erythroblasts after knockdown of Bag1, but they were severely delayed in their differentiation (Figure 4C-D). Bag1 functions as a nucleotide exchange factor for HSC70/HSP70, a chaperone that was reported to protect GATA1 from caspase-3-mediated cleavage during terminal erythroid differentiation.37 Furthermore, Bag1 links the protein surveillance chaperone system with the proteasome degradation,38 and a lack of Bag1 may enhance the unfolded protein response, which suppresses mRNA translation. This may be beneficial in a situation in which ribosome synthesis is restricted.
Mouse embryos lacking Csde1 die at approximately day E10.5 after gestation, suggesting a function for Csde1 in tissue development.25 Csde1 is an RNA-binding protein also known as Unr. In Drosophila, Unr is recruited by the sex lethal (SXL) protein to the 3′UTR of male-specific lethal 2 (msl-2) to inhibit translation.39 In mammalian cells, Csde1 was shown to bind the IRES of p58PITSLRE kinase and to enhance its translation.40 p58PITSLRE kinase is important for the G2-M transition of the cell cycle and is crucial for the completion of cytokinesis.41 Haploinsufficiency of Rpl24, a ribosomal protein that was not shown to be mutated in DBA, also impairs IRES-mediated translation and particularly the expression of p58PITSLRE.42 It will be interesting to determine whether this involves regulation of Csde1 expression or if haploinsufficiency of ribosomal proteins affects the translation of p58PITSLRE directly. The increase in multinuclear cells induced by the loss of Rps19 or Rpl11 (supplemental Figure 1C) indicates that mitosis, and more specifically cytokinesis of erythroblasts, is affected, similar to what was observed in a DBA mouse model expressing dominant-negative Rps19.43 It is possible that mitosis in such rapidly cycling cells relies more on proper IRES-driven translation of certain transcripts than in other cell types.
Although the knockdown and knockout models clearly demonstrate that Bag1 and Csde1 are required for erythropoiesis, the moderately reduced expression levels in DBA samples may not be sufficient for any single gene to cause anemia. Similarly, the brain defect observed in Bag1-deficient mice is not typical for DBA, and, for the same reason, heterozygous mice do not have a phenotype.21,44 Bag1 and Csde1 are only 2 examples from the list of translationally regulated genes. We propose that reduced translation of a set of transcripts, including Bag1 and Csde1, that share a common regulatory element contributes to a compound effect that results in the severe anemia of DBA.
In the present study, our patient cohort is too small and heterogeneous to draw conclusions about a relationship between impaired translation of Bag1 or Csde1 and the severity of the disease or the mutated ribosomal protein, although translation seems to be less affected in the 3 cultures that expanded relatively well. Moreover, the expression data in cultured DBA-derived erythroblasts are biased because we could only expand erythroblasts from 7 of 11 patients.
It has also been suggested that the severe anemia associated with DBA is caused by a p53-dependent mechanism, and that erythroblasts are extremely sensitive to activation of p53.9 A disbalance in ribogenesis enables quenching of Mdm2 by free Rpl5 or Rpl11, which reduces p53 degradation and activates p53-dependent pathways.45 Our data indicate that certain transcripts are specifically lost from polyribosomes in erythroblasts of DBA patients independently of p53 activation. However, we also show that Siva1 is lost from polysomes after Rps19 knockdown (from 70% to 30% polyribosome loading). Siva1 is a p53 target gene involved in negative feedback, and its reduced expression may enhance the activity of p53.46 Therefore, it seems likely that the overall DBA phenotype is caused by a combination of defective mRNA translation and p53 activation. This is supported by experiments in zebrafish also showing that p53-dependent and p53-independent mechanisms cooperate in the overall DBA phenotype caused by Rps19 knockdown.11,12
The recent finding that loss of RPS14 contributes to the phenotype of a subtype of myelodysplastic syndrome (MDS) characterized by loss of chromosome arm 5q47 raised the question of whether BAG1 and CSDE1 are aberrantly expressed in 5q− MDS. Because of the predominantly erythroid expression, however, BAG1 and CSDE1 protein expression was not detected in unfractionated BM, nor in the blasts present in MDS with or without a 5q aberration (data not shown). Therefore, it is difficult to envision how the loss of RPS14 and potential downstream targets such as BAG1 and CSDE1 contributes to the clonal expansion of blasts in MDS. Nevertheless, both DBA and 5q− MDS have a relatively low risk of leukemic transformation. Loss of RPS14 is likely to exert the specific erythroid phenotype of 5q− MDS, and at the same time, it may protect from leukemic transformation, similar to the mechanism through which haploinsufficiency of Rpl24 protects mice from myc-induced cancer.42
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank E. Bindels for valuable experimental suggestions and BM cell cDNA, C. Erpenlick-Verschuren for assistance with microarrays, A. Sanzsanz for the help with statistical analysis, M. Piwitzerova-Horvathova for the help with the patient material, P. van Strien and J. Haanstra for assistance with FACS, and E. van den Akker for critical reading of the manuscript.
This study was supported by grants from the European Community (Marie Curie RTN EUrythron grant MRTN-CT-2004-005499); the Ministry of Education, Czech Republic (MSM 6198959205); the Ministry of Health, Czech Republic (NT11059); and the German Research Foundation (DFG, SFB 581, TP B1).
Authorship
Contribution: R.H. performed most of the experiments; H.I. performed the Rpl11 array experiments and qRT-PCR array validations; R.S. and M.S. managed the Bag1 mice cohort; C.A.-S. derived the MEL-Csde1 cells; E.T. analyzed the array data; A.N. assisted with the bicistronic assays; R.C. and D.P. managed the patient-related work; and R.H., D.P., I.P.T., and M.v.L. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R.H. is European Molecular Biology Laboratory, Heidelberg, Germany. The current affiliation for H.I. is the Department of Immunology, Erasmus University Medical Center, Rotterdam, The Netherlands. The current affiliation for A.N. is the Department of Cell Biology, Sciences III, University of Geneva, Switzerland.
Correspondence: Marieke von Lindern, Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory AMC/UvA, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: m.vonlindern@sanquin.nl.


 and B
and B  ) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.
) Cumulative cell numbers are calculated from 3 independent experiments. (C) Cell size profiles, measured by a cell counter, from differentiating Bag1-deficient I/11 erythroblasts shown on the day of Epo induction (0h) and after 96 hours. Peaks at 10 μm indicate erythroblasts; peaks at 5 μm represent enucleated reticulocytes. (D) I/11 erythroblasts were transduced with lentiviral shRNA constructs as indicated. Cells were cultured for 3 days in proliferation conditions (top panels), followed by 4 days in differentiation conditions (bottom panels) before being harvested for cytospins and stained for hemoglobin (brown color) and histological dyes. Bars represent 25 μm.