We describe a novel clinical phenotype associating T- and B-cell lymphopenia, intermittent neutropenia, and atrial septal defects in 3 members of a consanguineous kindred. Their clinical histories included recurrent bacterial infections, viral infections, mucocutaneous candidiasis, cutaneous warts, and skin abscesses. Homozygosity mapping and candidate gene sequencing revealed a homozygous premature termination mutation in the gene STK4 (serine threonine kinase 4, formerly having the symbol MST1). STK4 is the human ortholog of Drosophila Hippo, the central constituent of a highly conserved pathway controlling cell growth and apoptosis. STK4-deficient lymphocytes and neutrophils exhibit enhanced loss of mitochondrial membrane potential and increased susceptibility to apoptosis. STK4 deficiency is a novel human primary immunodeficiency syndrome.
Introduction
Monogenic disorders of the human immune system have provided important insights into the function of host defense mechanisms.1 Despite remarkable progress in the field, many disorders remain poorly understood.2,3 Identifying genetic mutations in patients with immunodeficiency syndromes may reveal novel insights into basic mechanisms of the human immune system.
Here, we describe the first human patients with a biallelic mutation of serine threonine kinase 4 (STK4; MIM: 604965). STK4 (previously sometimes named MST1) was originally identified as a ubiquitously expressed kinase with structural homology to yeast Ste20.4,5 STK4 and STK3 (MST2; MIM: 605030) are the mammalian homologs of the Drosphila Hpo protein, the central constituent of the highly conserved HIPPO pathway controlling cell growth, apoptosis, and tumorigenesis.6 Mice lacking either Stk3 or Stk4 are viable, but those lacking both proteins are not. This indicates that each protein can substitute for the other in the most essential functions.7 When both Stk3 and Stk4 are conditionally deleted, however, their respective role as growth control regulators becomes manifest, exemplified by liver-specific double-knockout mice that develop massive hepatomegaly and hepatocellular carcinoma.8,9
STK4 has both proapoptotic and antiapoptotic functions. Earlier papers focused on the proapoptotic functions, and STK4 was described with the adjective “proapoptotic” in the title of a paper as recently as 2007.10 The strongest evidence that STK4 delivers proapoptotic signals is that STK4 is cleaved by caspases11,12 ; caspase activity is unambiguously proapoptotic. In resting conditions, STK4 is a cytoplasmic protein. In response to apoptotic stimuli, the 63-kDa full-length protein is cleaved by caspases and a 36-kDa N-terminal fragment translocates to the nucleus and phosphorylates histones,13,14 suggesting that STK4 plays a proapoptotic role. STK4 is also in a proapoptotic regulatory loop with JNK.15,–17 Finally, the interaction between RASSF1A and STK4 was shown to promote Fas-mediated apoptosis.18
There was also some evidence, before the generation of Stk4-deficient mice, that STK4 has antiapoptotic functions. For example, a study in Caenorhabditis elegans showed that phosphorylation of FOXO proteins by the STK4 ortholog DAF16 protects against cell death induced by oxidative stress. Furthermore, when DAF16 cannot perform the phosphorylation function, the life span of the worms is measurably reduced.19
Surprisingly, Stk4-deficient mice had progressive loss of T and B cells because of excessive apoptosis.20,–22 Thus, STK4 may also have a protective role maintaining cellular viability. STK4 phosphorylates transcription factors in the FOXO family, including FOXO1 and FOXO3, as part of a stress-response pathway.19,21 STK4 participates in several other pathways. Binding of NORE1A and RASSF1A to STK4 homodimers inhibits STK4 kinase activity.20,23 Binding of RAPL to STK4 is essential for lymphocytes to polarize and adhere24 and potentially to control proper egress from thymus.22
Our discovery of patients lacking STK4 allows a comparison between mice versus humans and highlights the physiologic role of the HIPPO pathway for the development of the immune and cardiac system.
Methods
Participants
Principal participants of the study were 8 related persons of Iranian ancestry. Blood and bone marrow samples were taken from healthy and affected family members and unrelated healthy persons. Biopsies of warts were taken from patients P2 and P3. Blood was taken from 100 unrelated Iranian controls and from members of 16 other consanguineous families with neutropenia for the purpose of sequencing STK4 or STK3.
Samples were taken on informed consent/assent, following current European regulations and the Declaration of Helsinki. The study was approved by the institutional review boards at Hannover and Isfahan Medical Schools. All patients were physically seen and examined at Hannover Medical School in 2010. Basic clinical information was retrieved from the patients' medical records.
Genotyping, linkage analysis, and gene sequencing
The samples from affected and healthy family members were genotyped using the Affymetrix 250k NspI SNP mapping array (GEO Platform GPL3718), following the same procedures recommended by Affymetrix, as done previously.25 Microsatellites were genotyped using published methods.26,–28 Exons and flanking intron-exon boundaries of candidate genes were amplified by PCR and sequenced with the use of ABI PRISM 3130 DNA Sequencer and DNA Sequencing Analysis Version 3 software (Applied Biosystems). To find homozygous, perfectly segregating intervals, we used the findhomoz software described previously.25 To compute LOD scores with SuperLink,28 we hypothesized that I-1, I-2, and II-1 share a common ancestor. Persons I-1 and I-2 are assumed to have a pair of great grandparents in common, and this pair is also assumed to be great-great grandparents of II-1. The assumption of common great grandparents to represent likely consanguinity has been used elsewhere.27,29 Other linkage parameters were set as in previous studies.26,27
Supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) shows the primers used to sequence the gene STK4. PCR conditions were as follows: 1 cycle initial denaturation 95°C for 5 minutes, followed by 30 cycles of 95°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute. A final elongation step of 72°C for 10 minutes ended the amplification.
Cells, antibodies, immunoblot, spectratyping, and flow cytometry
Protein extracts from PBMCs and EBV-immortalized B-cell lines from patients and healthy persons were separated by SDS-PAGE, blotted, and immunostained with antibodies to STK4 (Cell Signaling), FOXO3 (Cell Signaling), and GAPDH (Santa Cruz Biotechnology). Next, staining with HRP-conjugated secondary antibodies (BD Biosciences) was performed, blots were developed using a Chemiluminescence Kit (Pierce Chemical), and images were captured on a Chemidoc XRS Imaging System (Bio-Rad). Immunophenotyping of lymphocytes was performed using fluorochrome-conjugated antibodies as listed: anti-CD4–peridinin chlorophyll protein, anti-CD62L–allophycocyanin, anti-IgD–FITC, anti-CD21–PE (BD Biosciences); anti-CD8a–AlexaFluor-780, anti-CCR7–biotin, anti-CD38–allophycocyanin, anti-CD27–Biotin (eBioscience); anti-CD3–PeCy7, anti-CD19–PeCy7, anti-CD45RA–FITC, anti-CD45RO–PE (Beckman Coulter), anti-IgM–Pacific Blue (BioLegend). Cells stained with biotinylated antibodies were incubated with streptavidin-peridinin chlorophyll protein or eFluor450. PBMCs were acquired by a FACSCanto or BD LSRII (BD Biosciences), and data analysis was performed using FlowJo Version 9.4.10 software. TCRVβ spectratyping was performed as described previously26 with the exception that we used bone marrow mononuclear cells instead of PBMCs because of limitations of patient material.
Assessment of apoptosis and mitochondrial membrane potential of neutrophils and T lymphocytes
Neutrophils were isolated from peripheral blood by density gradient centrifugation. The purity was always more than 95%. Neutrophils were exposed to staurosporine (5μM; Sigma-Aldrich) at various time points and analyzed by FACS after staining with annexin-V (Invitrogen) and propidium iodide (Sigma-Aldrich). Cells were gated on intact neutrophils based on forward scatter and side scatter features. Dissipation of the mitochondrial membrane potential (MMP) was determined by FACS after loading the cells with valinomycin (100nM; Sigma-Aldrich) and JC-1 dye (3.5μM; Invitrogen).30
PBMCs isolated by gradient centrifugation over Ficoll Paque (GE Healthcare) were incubated with anti-Fas monoclonal antibody (5 μg/mL) or staurosporine (5μM) for various time points and analyzed by FACS after staining with propidium iodide and CD3 antibody (BD Biosciences). Loss of MMP was determined by FACS after loading the PBMCs with valinomycin (100nM; Sigma-Aldrich) and staining with CMXRos (Invitrogen) and gating on CD3+ population.
Results
Clinical phenotype
We investigated 3 patients from a consanguineous Iranian family presenting with a primary immunodeficiency, including bacterial infections, viral infections, mucocutaneous candidiasis, and cutaneous warts. Female patient P1, born in 1986, had pneumonia at age 2 years and subsequently recurrent upper and lower respiratory tract infections. Her brother P2, born in 1990, presented at age 10 years with neutropenia and recurrent episodes of fever and upper respiratory tract infections. Two siblings of P1 and P2 died in their first year of life because of septicemia. Female patient P3 was born in 2001 and presented with recurrent sinusitis and rhinitis at age 4 years. She had at least 2 episodes of staphylococcal pneumonia. All 3 patients had histories of recurrent skin abscesses.
All patients showed continuously or intermittently decreased numbers of peripheral neutrophil granulocytes (Table 1; supplemental Table 2); however, in contrast to severe congenital neutropenia secondary to mutations in HAX1 or ELANE, maturation of neutrophils in the bone marrow appeared normal in all 3 patients (Figure 1A; supplemental Figure 1). Neutropenia was identified also in periods when the patients did not have infections, suggesting that low counts of neutrophil granulocytes were not induced by infections. No growth defects or dysmorphic features were noted. The patients had unremarkable heights and weights.
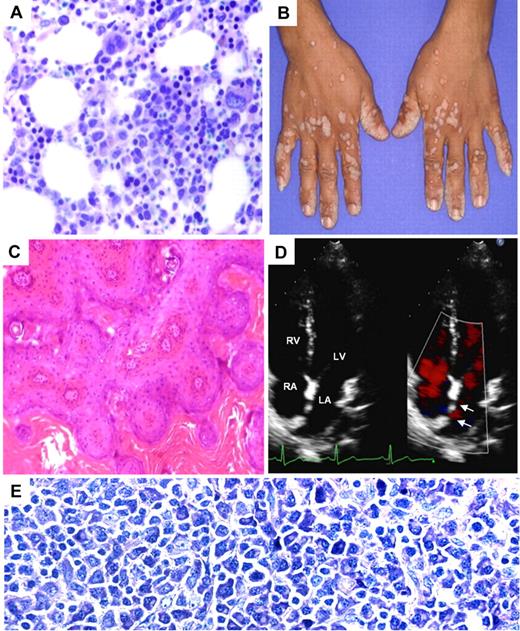
Clinical phenotype of STK4-deficient patients. (A) H&E-stained bone marrow biopsy shows mild increase in cellularity and full maturation of neutrophil granulocytes. (B) The photograph shows disseminated warts infections in P2. (C) H&E-stained sections from wart biopsy shows orthokeratotic verrucosis and epithelial hyperplasia (P2; original magnification ×100). (D) Echocardiogram (P1) showing atrial septal defect type II (arrow) and left-right shunt. RA indicates right atrium; LA, left atrium; RV, right ventricle; and LV, left ventricle. (E) Giemsa staining of the enlarged inguinal lymph node of P3, showing accumulation of plasmacytoid differentiated cells (original magnification ×400).
Clinical phenotype of STK4-deficient patients. (A) H&E-stained bone marrow biopsy shows mild increase in cellularity and full maturation of neutrophil granulocytes. (B) The photograph shows disseminated warts infections in P2. (C) H&E-stained sections from wart biopsy shows orthokeratotic verrucosis and epithelial hyperplasia (P2; original magnification ×100). (D) Echocardiogram (P1) showing atrial septal defect type II (arrow) and left-right shunt. RA indicates right atrium; LA, left atrium; RV, right ventricle; and LV, left ventricle. (E) Giemsa staining of the enlarged inguinal lymph node of P3, showing accumulation of plasmacytoid differentiated cells (original magnification ×400).
An immunologic investigation of all 3 patients in 2010 (Table 1) revealed lymphopenia with a paucity of T and B cells in P1 and P2. P3 had low numbers of CD4+ T cells and B cells but elevated counts of CD8+ T cells. At the time of the study, P3 presented with generalized EBV-associated lymphadenopathy, which may explain the high levels of CD8+ T cells. Patient 3 had episodes of lymphopenia in her clinical records (596 CD3+ cells/μL, 187 CD19+ cells/μL at age 8; supplemental Figure 2).
Consistent with defective adaptive immunity, all patients have had viral infections. P1 (not shown) and P2 (Figure 1B) had cutaneous warts. Histologic analysis of warts for P2 showed orthokeratosis and verrucous epithelial hyperplasia (Figure 1C) caused by human papillomavirus 57 (HPV 57) and HPV 84. Patient 3 had flesh-colored cutaneous molluscum contagiosum infection and cutaneous papillomatosis associated with multiple types of HPV (HPV 71, HPV 3, and HPV 25). All 3 patients had recurrent mouth ulcers consistent with the clinical diagnosis of herpes simplex virus. The herpes simplex virus serology results for IgG were 13 022 U/L, 17 046 U/L, and 22 971 U/L for P1, P2, and P3, respectively. No signs of active infection with varicella-zoster virus, measles, mumps, or HIV were observed in any of the 3 patients. However, the patients did have detectable antibody titers against herpes simplex virus, varicella-zoster virus, EBV, measles, tetanus, diphtheria, and mumps that would be consistent with either vaccination or an earlier infection. The titers against diphtheria, tetanus, and pertussis (DTP) were measured when the patients were evaluated in Germany in 2010. Based on their clinical history information, the DTP vaccinations were given 4 (P1), 5 (P2), and 6 (P3) years before the DTP titer measurement.
P3 had abnormal heart sounds detected by physical examination. Systematic echocardiography of all 3 patients in 2010 identified structural cardiac abnormalities, including atrial septal defect type II (P1; Figure 1D), patent foramen ovale (P2), and patent foramen ovale associated with mitral, tricuspid, and pulmonary insufficiency (P3). These cardiac aberrations have not caused clinical symptoms.
Genetic analysis revealed loss-of-function mutation in STK4
By gene sequencing, we excluded the possibility that these patients had an unusual form of a known primary immunodeficiency, such as WHIM syndrome caused by mutations in CXCR4,31 G6PC3 deficiency,26 or severe congenital neutropenia caused by mutations in either ELANE or HAX1.30,32 None of these genes was mutated. We performed a SNP-based genome-wide homozygosity mapping study, which we refined using microsatellite markers according to published methods.25 We identified a single region in which markers segregate perfectly with disease (Figure 2A). This was on chromosome 20 and spans at least 41.2 to 43.2 Mbp (build 36/hg18) and at most 41.2 to 43.6 Mbp. We genotyped microsatellites D20S96 and D20S481 to confirm the genetic linkage and to clarify the telomeric boundary. Multipoint analysis yielded a LOD score of 4.3.
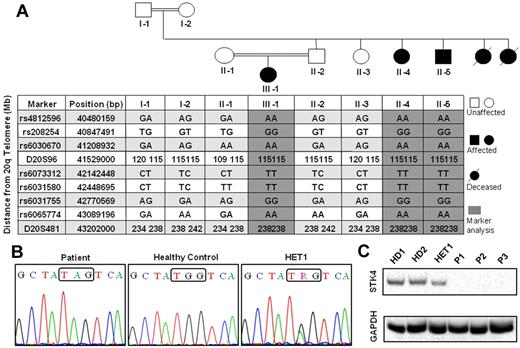
Linkage analysis of index family, STK4 mutation, and absence of STK4 protein expression in the patients. (A) Pedigree of the family with SNP and microsatellite markers on chromosome 20. Gray shading represents the homozygous interval. Nomenclature of the persons in the text is as follows: II-1 indicates HET2; II-2, HET1; II-4, P1; II-5, P2; and III-1, P3. (B) Sanger sequencing of STK4 shows a nucleotide substitution G/A in exon 7, which leads to premature stop codon mutation. (C) Detection of STK4 protein in PBMCs by Western blot. All 3 STK4-deficient patients do not show any protein expression.
Linkage analysis of index family, STK4 mutation, and absence of STK4 protein expression in the patients. (A) Pedigree of the family with SNP and microsatellite markers on chromosome 20. Gray shading represents the homozygous interval. Nomenclature of the persons in the text is as follows: II-1 indicates HET2; II-2, HET1; II-4, P1; II-5, P2; and III-1, P3. (B) Sanger sequencing of STK4 shows a nucleotide substitution G/A in exon 7, which leads to premature stop codon mutation. (C) Detection of STK4 protein in PBMCs by Western blot. All 3 STK4-deficient patients do not show any protein expression.
In 2008, before Stk4-deficient mice had been described, we sequenced 12 of the 46 genes in the maximal linkage interval (supplemental Table 3), prioritizing genes highly expressed in the hematopoietic system or having a role in apoptosis. STK4, located in the narrowest possible linkage interval, was a plausible candidate gene. All 3 patients had a homozygous stop codon mutation in exon 7 of the gene STK4 (c.G750A, p.W250X). Parents and healthy siblings were heterozygous for the mutation, consistent with autosomal recessive inheritance (Figure 2B). We sequenced the entire exon 7 in 100 healthy Iranian controls; no control carried any sequence change in this exon. We sequenced STK4 in 10 unrelated patients with consanguineous parents, 6 with neutropenia and markers segregating perfectly around STK4 and 4 with lymphopenia and neutropenia. We have not yet identified any other patients with a mutation in STK4. Similarly, we were unable to discover any mutations in STK3 in 6 unrelated patients born to consanguineous parents who had markers segregating perfectly around STK3.
To test whether the mutant STK4 protein is expressed, we performed Western blots using an antibody that recognizes an N-terminal epitope of STK4. Whereas PBMCs from healthy controls showed expression of STK4, no expression was seen in cells from patients with a homozygous W250X mutation (Figure 2C). Heterozygous carriers had intermediate levels of STK4 abundance. We infer that the premature termination abrogates STK4 translation, via nonsense-mediated decay of the mutant mRNA. Originally, STK4 was not the strongest candidate gene in the linkage interval because of a report that STK4 is not expressed in neutrophils.33 However, our Western blot results from isolated neutrophil granulocytes (supplemental Figure 3) and the functional data later within the “Discussion” prove that STK4 is expressed in neutrophils. In light of the previous negative report,33 the weakness of the Western blot is not surprising.
Immunologic analysis in STK4-deficient patients
We performed immune assays for all 3 patients (Table 1; supplemental Figure 2). Consistent with the phenotype of Stk4-deficient mice,20 all patients showed a reduced fraction of CD45RA+CD45RO− naive T cells (Figure 3A). Additional experiments to determine central and effector memory T cells show that CD62L+ CCR7+ cells T cells, also named central memory T cells, are decreased in STK-deficient patients. In contrast, the CD62L−CCR7− population of effector memory T cells appears less affected (Figure 3B). Molecular spectratyping on T-cell receptor-Vβ subclasses shows that the pseudo-Gaussian distribution of almost all Vβ subclasses seen in a healthy donor is disturbed in STK4-deficient patients (supplemental Figure 4).
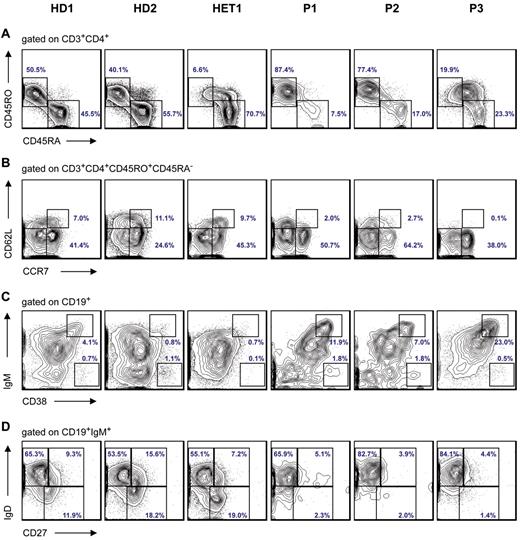
Immunophenotyping results of T and B cells in STK4-deficient patients. (A) Flow cytometric staining of peripheral CD3+CD4+ T cells for differentiation markers reveals relative increase of CD45RA−CD45RO+ memory T cells and decrease of CD45RA+CD45RO− naive T cells in PBMCs isolated from patients (P1, P2, and P3) compared with HD (healthy donors) and heterozygous. (B) Based on the expression of CD62L and CCR7, memory T cells were further divided into effector memory (CD45RO+CCR7−CD62L−) and central memory (CD45RO+CCR7+CD62L+) subsets. (C) The circulating B-cell pool was characterized by CD19 staining and further divided into B-cell subtypes. The immunophenotyping shows an increased fraction of transitional B cells (CD38++IgMhigh; D) and also reduction of marginal zone B cells (IgD+IgM+CD27+) and switched memory B cells (IgD−IgM−CD27+). The normal range for switched memory B cells in children 6 to 10 years of age is 5.2% to 12.1%; and for adults 19 to 25 years of age, it is 7.2% to 12.7%.34
Immunophenotyping results of T and B cells in STK4-deficient patients. (A) Flow cytometric staining of peripheral CD3+CD4+ T cells for differentiation markers reveals relative increase of CD45RA−CD45RO+ memory T cells and decrease of CD45RA+CD45RO− naive T cells in PBMCs isolated from patients (P1, P2, and P3) compared with HD (healthy donors) and heterozygous. (B) Based on the expression of CD62L and CCR7, memory T cells were further divided into effector memory (CD45RO+CCR7−CD62L−) and central memory (CD45RO+CCR7+CD62L+) subsets. (C) The circulating B-cell pool was characterized by CD19 staining and further divided into B-cell subtypes. The immunophenotyping shows an increased fraction of transitional B cells (CD38++IgMhigh; D) and also reduction of marginal zone B cells (IgD+IgM+CD27+) and switched memory B cells (IgD−IgM−CD27+). The normal range for switched memory B cells in children 6 to 10 years of age is 5.2% to 12.1%; and for adults 19 to 25 years of age, it is 7.2% to 12.7%.34
The circulating B-cell pool was characterized by CD19 staining and further distinguished as naive B cells (IgD+IgM+CD27−), marginal zone B cells (IgD+IgM+CD27+), switched memory B cells (IgD−IgM−CD27+), activated CD21 lowCD38low B cells, transitional B cells (CD38++IgMhigh), and CD38+++IgM− class-switched plasmablasts. All patients had decreased numbers of CD19+ cells, an increased fraction of transitional B cells (CD38++IgMhigh; Figure 3C), and a reduction of marginal zone B cells (IgD+IgM+CD27+) and switched memory B cells (IgD−IgM−CD27+; Figure 3D). Despite the peripheral B-cell lymphopenia, all patients had evidence of hypergammaglobulinemia. P1 and P3 had increased levels of IgG; P2 and P3 had increased levels of IgA. IgM levels were decreased in P1 and P2. All 3 patients showed elevated levels of IgE (Table 1). All patients were immunized and produced protective levels of specific antibodies to diphtheria, tetanus, and polio. Interestingly, all patients had measurable levels of autoantibodies (red blood cells, thyroglobulin; Table 1), possibly because of unrestricted plasma cell expansion.
Calcium flux of isolated neutrophils on stimulation with formyl-methionyl-leucyl-phenylalanine was tested at 2 different time points. There was not any meaningful difference between the patient and healthy controls (supplemental Figure 5).
A lymph node biopsy in P3 (performed in view of chronic lymphadenopathy) showed an EBV-associated B-lymphoproliferative disorder with plasmacytoid differentiation, κ-light chain restriction, and monoclonal immunoglobulin gene rearrangement (Figure 1E; supplemental Figure 6A-B). Furthermore EBER in situ hybridization proved EBV infection of infiltrates (supplemental Figure 6C-D). Phenotypically, the EBV-associated lymphoproliferative disorder resembled lymphoplasmacytic lymphoma. EBV serology on P3 showed EBV-IgG positive (1570 U/mL) and EA-C-IgG positive. Furthermore, EBV-DNA PCR of lymph node biopsy showed 12 copies per cell. For P1, EBV serology showed IgG 998 U/mL and IgM negative; for P2, EBV serology showed both IgG and IgM to be negative.
Increased apoptosis in STK-4 deficient T cells and neutrophil granulocytes
Stk4-deficient mouse T lymphocytes are highly susceptible to apoptosis,21 yet no phenotype of murine Stk4-deficient neutrophil granulocytes has been reported. Purified peripheral T cells were stimulated with anti-Fas monoclonal antibody, and apoptosis was measured by staining with annexin-V and propidium iodide. Furthermore, purified peripheral neutrophils were exposed to staurosporine, and apoptosis was measured by flow cytometry. As shown in Figure 4A-B, STK4-deficient cells exhibited a higher degree of apoptosis than did cells from control or heterozygous persons. Similar findings were observed when T cells were exposed to staurosporine (supplemental Figure 7A).
Functional studies of T lymphocytes and neutrophils. (A) Time course of induced apoptosis in STK4-deficient T cells on exposure to anti-Fas, which was measured by staining with annexin-V and propidium iodide. STK4-deficient T cells exhibited a significantly higher degree of apoptosis than did cells from control or heterozygous persons. (B) Time course of induced apoptosis in STK4 neutrophils on exposure to staurosporine. Patients' neutrophils showed increased apoptosis compared with healthy persons. (C) Visualization of gradual loss of mitochondrial membrane potential evidenced by fading CMXROS fluorescence intensity in T cells. Note that patients P1 and P2 have accelerated loss of Δψm compared with healthy control cells. (D) Enhanced loss of mitochondrial membrane potential Δψm in STK4-deficient neutrophil granulocytes, which is measured by fading JC-1 dye fluorescence intensity. (E) Decreased FOXO3 expression in PBMCs of STK4-deficient patients revealed by Western blot. Both the apoptosis and mitochondrial membrane potential assays were done twice, on independently purified cells.
Functional studies of T lymphocytes and neutrophils. (A) Time course of induced apoptosis in STK4-deficient T cells on exposure to anti-Fas, which was measured by staining with annexin-V and propidium iodide. STK4-deficient T cells exhibited a significantly higher degree of apoptosis than did cells from control or heterozygous persons. (B) Time course of induced apoptosis in STK4 neutrophils on exposure to staurosporine. Patients' neutrophils showed increased apoptosis compared with healthy persons. (C) Visualization of gradual loss of mitochondrial membrane potential evidenced by fading CMXROS fluorescence intensity in T cells. Note that patients P1 and P2 have accelerated loss of Δψm compared with healthy control cells. (D) Enhanced loss of mitochondrial membrane potential Δψm in STK4-deficient neutrophil granulocytes, which is measured by fading JC-1 dye fluorescence intensity. (E) Decreased FOXO3 expression in PBMCs of STK4-deficient patients revealed by Western blot. Both the apoptosis and mitochondrial membrane potential assays were done twice, on independently purified cells.
We also tried to examine in vitro proliferation of STK4-deficient T cells. Because of rapid death of STK4-deficient T cells in vitro, we cannot provide conclusive data on in vitro proliferation in response to mitogens or antigens. Supplemental Figure 7B shows rapid onset of apoptosis in T cells after stimulation with anti-CD3, anti-CD28, and recombinant human IL-2.
In view of a recent report implicating increased loss of the mitochondrial transmembrane potential (Δψm) in Stk4−/− peripheral T cells,21 we next visualized loss of Δψm in primary patients' cells by flow cytometry. We obtained peripheral blood T cells and neutrophils from patients and healthy persons. T cells were loaded with chloromethyl-X-rosamina (H2-CMX-Ros), a cationic lipophilic fluorochrome sensitive to changes in Δψm.35 Neutrophils were assayed using JC-1 dye, also sensitive to changes in Δψm. On exposure to valinomycin, STK4-deficient T cells and neutrophil granulocytes showed a rapid loss of Δψm, whereas cells from heterozygous and STK4+/+ persons showed a much slower loss, suggesting that increased apoptosis of STK4-deficient leukocytes is at least partly the result of increased dissipation of Δψm (Figure 4C-D).
FoxO3a, a direct downstream target of Stk4, has previously been shown to protect against apoptosis in Stk4-deficient mice.21 We further hypothesized that STK4-deficient T cells may display increased sensitivity to reactive oxygen species. Indeed, human STK4-deficient T cells had decreased levels of FoxO3a, as measured by Western blot assays (Figure 4E). The mRNA expression level of FoxO3a from isolated PBMCs of the patients was also decreased compared with healthy controls (supplemental Figure 8). Thus, STK4 may protect against increased oxidative stress and susceptibility to apoptosis via FoxO proteins.
Discussion
Human STK4 deficiency causes a primary immunodeficiency syndrome affecting T cells, B cells, and possibly neutrophil granulocytes. Several features, such as lymphopenia and increased susceptibility to apoptosis, recapitulate findings in Stk4-deficient mice.20,–22 However, our analysis of human patients has highlighted certain novel phenotypic characteristics deserving further investigations. All 3 patients showed a relative increase in transitional B cells and evidence of hypergammaglobulinemia, associated with increased titers of autoantibodies. One patient had EBV-associated lymphoproliferative disease. It remains to be shown whether unrestricted B-cell expansion is the result of defective T-cell responses or intrinsic mechanisms in STK4-deficient B cells.
Intermittent neutropenia was observed in all 3 patients, although neutropenia has not been reported in Stk4-deficient mice. Neutrophils from patients exhibited more apoptosis than did neutrophils from healthy controls or heterozygous persons. Species-dependent mechanisms controlling viability of neutrophil granulocytes have been noted in other comparative studies.36 Patients did not exhibit a defect in neutrophil production; bone marrow samples were studied in both Iran and Germany, and there was no evidence of a block in myeloid differentiation. Two of 3 patients (P2 and P3) had no evidence of antineutrophil antibodies, whereas patient P1 had borderline positive test results. These findings do not allow us to explain decreased neutrophil counts by postulating an autoimmune mechanism, yet they are compatible with such a mechanism.
Our attempts to discover additional patients were not successful. The group of Geneviève de St Basile undertook a genetic linkage study in 2 unrelated Turkish families and has identified other loss-of-function mutations in STK4 (see Nehme et al37 ). Future studies are needed to determine the frequency of STK4 mutations in patients with primary immunodeficiency disorders.
Proof of causality would have been strengthened by functional reconstitution assays. However, STK4-deficient T cells could not be maintained in vitro (supplemental Figure 7B) because of premature apoptosis. Similarly, our attempts to generate T cells from CD34+ cells in vitro failed because of premature cell death. In light of the various roles of STK4 in apoptosis reviewed in the “Introduction,” the difficulties in maintaining STK4-deficient cells alive are not entirely surprising. Myeloid cells derived from STK4-deficient progenitors in vitro did not show a discernible phenotype, potentially secondary to the antiapoptotic effect of the high levels of cytokine exposure needed for in vitro differentiation.
In addition to immunologic defects, all 3 STK4-deficient patients had structural cardiac aberrations. Interestingly, transgenic mice with cardiac-specific overexpression of Stk4 (Tg-Stk4) developed dilated cardiomyopathy secondary to increased apoptosis of myocytes,38 but no cardiac phenotype has been reported in Stk4-deficient mice.20,21 Stk4 is activated by Rassf1A in the heart, promoting apoptosis in cardiac cells and inhibiting cardiac fibroblast proliferation, thus controlling cardiac remodeling,23 and mutations in other murine genes in the STK4/Hippo pathway also lead to heart defects.39
In conclusion, we describe a novel primary immunodeficiency syndrome caused by a homozygous nonsense mutation in STK4, the ortholog to Drosophila Hpo. STK4 is critical for maintenance of lymphocytes and control of unrestricted EBV-induced lymphoproliferation.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all patients and healthy members of the family, the medical staff at Hannover Medical School and Isfahan University of Medical Sciences for expert support, Andreas Krüger and Marcin Lyszkiewicz for help with T-cell differentiation experiments, Ursula Berka for help in performing neutrophil functional studies, and Judy Levin for administrative support.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB900, KFO250, and Gottfried-Wilhelm-Leibniz program), the Care-for-Rare Foundation, the Deutsche José Carreras Leukämie-Stiftung, the European Union commission Marie-Curie (grants MEXT-CT-2006-042316 and FP7/2007-2013), EURO-PADnet (grant HEALTH-F2-2008-201549), and in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. H.A. is a graduated member of the international MD-PhD program at Hannover Medical School.
National Institutes of Health
Authorship
Contribution: H.A. provided patients' samples, analyzed genetic data, sequenced genes to identify the STK4 mutation, performed Western blots, performed functional studies, and helped write the manuscript; G.A. performed functional studies and immunophenotyping; D.K. performed the FACS experiments; J.D. performed the Vβ spectratyping; R.B. cared for the patients during their stay in Hannover and helped gather all the diagnostic data; A.A.S. and E.M.G. performed the genetic linkage analysis and helped write the manuscript; A.S. performed retroviral gene transfer; H.H.K. analyzed the histopathologic findings of the patients; D.P. performed SNP genotyping; K.R.E. did microsatellite genotyping; N.R. introduced the index family to us; B.G. provided laboratory resources and supervised K.R.E.; S.L. performed echocardiography of all patients; R.S. performed clinical evaluations and treated the patients in Iran, arranged for laboratory studies in Germany, and ascertained their samples for this study; and C.K. directed the study, evaluated the patients, designed experiments, supervised H.A. and G.A., and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christoph Klein, University Children's Hospital Munich, Dr von Haunersches Kinderspital, Lindwurmstraße 4, D-80337 Munich, Germany; e-mail: christoph.klein@med.uni-muenchen.de.