Abstract
The protein tyrosine phosphatase CD45, encoded by the PTPRC gene, is well known as a regulator of B- and T-cell receptor signaling. In addition, CD45 negatively regulates JAK family kinases downstream of cytokine receptors. Here, we report the presence of CD45 inactivating mutations in T-cell acute lymphoblastic leukemia. Loss-of-function mutations of CD45 were detected in combination with activating mutations in IL-7R, JAK1, or LCK, and down-regulation of CD45 expression caused increased signaling downstream of these oncoproteins. Furthermore, we demonstrate that down-regulation of CD45 expression sensitizes T cells to cytokine stimulation, as observed by increased JAK/STAT signaling, whereas overexpression of CD45 decreases cytokine-induced signaling. Taken together, our data identify a tumor suppressor role for CD45 in T-cell acute lymphoblastic leukemia.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy characterized by the accumulation of undifferentiated thymocytes that have acquired multiple genomic aberrations affecting critical transcriptional and signaling pathways.1,2 T-ALL is also frequently characterized by the expression of constitutively activated tyrosine kinases, such as ABL1, LCK, JAK1, and JAK3.3-7 Recently identified mutations in the IL-7 receptor α (IL-7R) and deletions of the tyrosine phosphatase PTPN2 were also reported to affect tyrosine kinase signaling.8-10 The genetic and functional data presented in this work now identify the tyrosine phosphatase CD45 as a new tumor suppressor gene in T-ALL. CD45 is a transmembrane protein that is abundantly present on the surface of all nucleated hematopoietic cells. CD45 is encoded by the PTPRC gene and is known to regulate phosphorylation of SRC and JAK family kinases.11-13
Methods
Cell culture
HEK293T and human T-ALL cell lines were cultured in RPMI 1640 medium supplemented with FBS. MOHITO cells were cultured and transduced as described previously.14 For dose-response curves, T-ALL cell lines were seeded out in triplicate in 24-well plates at a density of 5 × 105 cells/mL and incubated for 48 hours with the JAK family kinase inhibitor INCB018424 (Chemietek). Viable cell numbers were determined using CellTiter 96 AQueous One Solution (Promega) and a Victor X4 plate reader (PerkinElmer Life and Analytical Sciences).
Patient samples and sequence analysis
Patient genomic DNA was collected at various institutions at time of diagnosis and remission. All samples were obtained according to the guidelines of the local ethics committees at KU Leuven, and informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. All coding exons of PTPRC as well as exon 6 of IL-7R were amplified and PCR products were directly sequenced.
Constructs
The open reading frames of wild-type and mutant CD45R0 were cloned into pMSCV-Puro (Clontech). Retroviral vectors expressing nonsilencing or CD45 targeting shRNAs were obtained by cloning short hairpin RNA sequences into a pMSCV-GFP construct containing a mir30 flanking cassette. shRNA sequences: CTCGCTTGGGCGAGAGTAA (shControl) and AGCAGATGATATTCCAAAGAAA (shCD45).
Western blotting
The following antibodies were used: anti-CD45 (clone 69; BD Biosciences); anti–phospho-JAK1 (Tyr1022/1023), anti-STAT5 (clone L-20; Santa Cruz Biotechnology); anti-JAK1 (clone73; Millipore); anti–phospho-STAT5 (Tyr694), anti–phospho-STAT3 (Tyr705), anti-STAT3 (79D7; Cell Signaling); anti–beta actin (Sigma-Aldrich).
Immunocomplex phosphatase activity assay
DiFMUP immunocomplex phosphatase activity assay15 was performed as described earlier.16 Conversion of DiFMUP into DiFMU was monitored using a Victor X4 plate reader. Initial conversion rates were calculated from the slope of the linear curve of fluorescence versus time for each substrate concentration.
Results and discussion
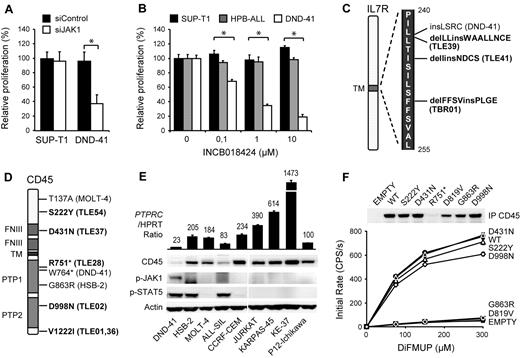
Protein tyrosine kinases are an important family of oncogenes that are frequently mutated in cancer. As their kinase activity is required for proliferation and survival of the cancer cells, tyrosine kinases have been of major interest as therapeutic targets.17,18 With the aim of identifying new therapeutic kinase targets in T-ALL, we performed RNAi screens in T-ALL cell lines with a tyrosine kinase-focused siRNA library. We electroporated 9 T-ALL cell lines (ALL-SIL, HSB-2, HPB-ALL, MOLT-14, KE-37, SUP-T1, DND-41, P12-ichikawa, and TALL-1) with our siRNA library, and searched for siRNAs that significantly affected proliferation and survival of the cells. As expected, we identified ABL1 (NUP214-ABL1) as an essential kinase in ALL-SIL cells,3 and LCK as an essential kinase in HSB-2 cells,5 2 cell lines known to depend on these oncogenic kinases (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, we identified tyrosine kinases that affected the proliferation and survival of several other T-ALL cell lines, the strongest hit being JAK1 in DND-41 cells (supplemental Figure 2). Using an independent JAK1 targeting siRNA as well as a JAK kinase family specific inhibitor, we confirmed that JAK1 kinase activity was strictly required for the proliferation of DND-41 cells (Figure 1A-B). Western blot analysis of DND-41 cells confirmed constitutive phosphorylation of JAK1 and its downstream target STAT5 (Figure 1E; supplemental Figure 3).
Identification of CD45 mutations in T-ALL. (A) DND-41 is sensitive to JAK1 knockdown. SUP-T1 and DND-41 cells were electroporated with a JAK1 targeting or scrambled control siRNA. Cell proliferation was normalized to scrambled control. The average ± SEM of 3 repeats is shown. *Statistical significance. (B) DND-41 is sensitive to JAK inhibition. SUPT-1, HPB-ALL, and DND-41 cells were treated with increasing concentrations of JAK inhibitor INCB018424. Cell proliferation was normalized to DMSO-treated cells. The average ± SEM of 3 repeats is shown. *Statistical significance. (C) IL-7R mutations identified in a sequencing screen in T-ALL patients (bold) and cell lines. (D) PTPRC nonsense and mis-sense variants identified in a sequencing screen in T-ALL patients (bold) and cell lines. (E) Western blot and quantitative PCR analysis of 9 T-ALL cell lines showing low CD45 expression and constitutive JAK1/STAT5 autophosphorylation in DND-41. PTPRC mRNA values are relative to hypoxanthine phosphoribosyl transferase and normalized to P12-Ichikawa. (F) Phosphatase activity of CD45 mis-sense variants. Beads coupled to wild-type and variant CD45 were incubated with increasing concentrations of DiFMUP, and initial rates of conversion to DiFMU were determined for each variant. The average ± SEM of 3 repeats is shown. Equal loading was assayed by Western blot analysis. D819V indicates phosphatase-dead CD45 mutant.
Identification of CD45 mutations in T-ALL. (A) DND-41 is sensitive to JAK1 knockdown. SUP-T1 and DND-41 cells were electroporated with a JAK1 targeting or scrambled control siRNA. Cell proliferation was normalized to scrambled control. The average ± SEM of 3 repeats is shown. *Statistical significance. (B) DND-41 is sensitive to JAK inhibition. SUPT-1, HPB-ALL, and DND-41 cells were treated with increasing concentrations of JAK inhibitor INCB018424. Cell proliferation was normalized to DMSO-treated cells. The average ± SEM of 3 repeats is shown. *Statistical significance. (C) IL-7R mutations identified in a sequencing screen in T-ALL patients (bold) and cell lines. (D) PTPRC nonsense and mis-sense variants identified in a sequencing screen in T-ALL patients (bold) and cell lines. (E) Western blot and quantitative PCR analysis of 9 T-ALL cell lines showing low CD45 expression and constitutive JAK1/STAT5 autophosphorylation in DND-41. PTPRC mRNA values are relative to hypoxanthine phosphoribosyl transferase and normalized to P12-Ichikawa. (F) Phosphatase activity of CD45 mis-sense variants. Beads coupled to wild-type and variant CD45 were incubated with increasing concentrations of DiFMUP, and initial rates of conversion to DiFMU were determined for each variant. The average ± SEM of 3 repeats is shown. Equal loading was assayed by Western blot analysis. D819V indicates phosphatase-dead CD45 mutant.
Although activating mutations in JAK1 have been reported in T-ALL,6,19,20 no such mutations are present in DND-41.21 We therefore sequenced all other JAK kinases as well as JAK1-associated cytokine receptors and negative regulators to determine the cause of constitutive JAK/STAT signaling in DND-41. We detected 2 mutations that could contribute to JAK1 activation. The first mutation was a heterozygous 12-nucleotide insertion in the IL-7R gene (IL-7R p.L242_L243insLSRC; Figure 1C; supplemental Figure 4). Similar mutations were recently reported in approximately 10% of T-ALL cases and were shown to be gain-of-function mutations activating the JAK/STAT pathway.8,10 Our data therefore confirm the critical role of JAK1 downstream of IL-7R mutants and identify DND-41 as a model system for their further study. In addition to the IL-7R mutation, we also detected a loss-of-function mutation W764* in the PTPRC gene, encoding the tyrosine phosphatase CD45 (Figure 1D; supplemental Figure 5). In agreement with this, we observed low expression of CD45 in DND41 cells as a result of the degradation of the W764* nonsense transcript (Figure 1E; supplemental Figure 6). These data suggested that CD45 could function as a tumor suppressor gene in T-ALL.
Next, we sequenced exon 6 of IL-7R and all coding exons of PTPRC in 12 additional T-ALL cell lines and in 65 T-ALL diagnostic patient samples. We identified 3 novel insertions and deletions in IL-7R (Figure 1C; supplemental Figure 4). Furthermore, we identified several PTPRC mutations that were likely to be pathogenic, as well as variations that were likely to be rare SNPs (Figure 1D; supplemental Figures 5 and 7; supplemental Table 1). In the HSB-2 cell line, we identified a G863R mis-sense mutation in the phosphatase domain and confirmed that this mutation causes loss of CD45 phosphatase activity (Figure 1F). In T-ALL patient TLE-28, we identified another inactivating nonsense mutation R751* similar to the W764* in DND-41.
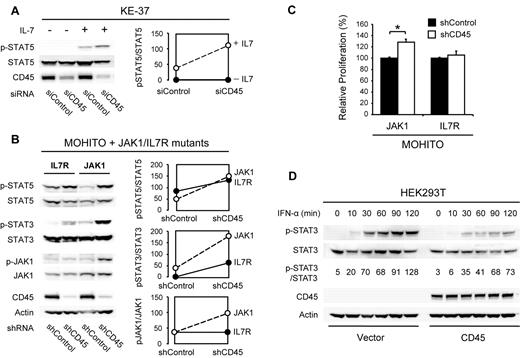
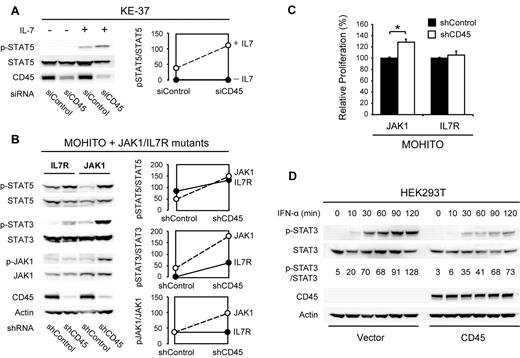
To determine the functional consequences of reduced CD45 function in T cells, we performed RNAi-mediated knockdown studies of CD45 in the human T-ALL cell line KE-37 and in the cytokine dependent mouse T-cell line MOHITO,22 which both have normal CD45 expression levels. In KE-37 cells, knockdown of CD45 caused increased sensitivity to cytokine stimulation, as shown by increased JAK/STAT pathway activity (Figure 2A). We then investigated whether loss of CD45 could also potentiate the effect of JAK1 or IL-7R activating mutations. Knockdown of CD45 indeed caused an increase in JAK/STAT pathway activity in MOHITO cells expressing mutant JAK1 and, to a lesser extent, mutant IL-7R (Figure 2B). In agreement with this, knockdown of CD45 also caused increased proliferation of MOHITO cells expressing mutant JAK1 (Figure 2C). On the other hand, overexpression of wild-type CD45 in HEK293T cells yielded a reduced sensitivity of the JAK/STAT signaling pathway to stimulation with interferon α (Figure 2D).
Effects of CD45 knockdown and overexpression. (A) Knockdown of CD45 increases sensitivity of the human T-ALL cell line KE-37 to IL-7 stimulation. KE-37 cells were electroporated with a CD45 targeting or control siRNA. At 72 hours after electroporation, a fraction of the cells were stimulated with 10 ng/mL IL-7 for 10 minutes. Phosphorylation of STAT5 was assayed by Western blot (left) and quantified (right). (B) Knockdown of CD45 increases JAK/STAT signaling in MOHITO cells transformed by JAK1 or IL-7R gain-of-function mutants. MOHITO cells transformed by activating JAK1 or IL-7R mutants were transduced with a CD45 targeting or control shRNA. Phosphorylation of JAK1 and STAT5 was assayed by Western blot (left) and quantified (right). (C) Knockdown of CD45 increases proliferation of MOHITO cells transformed by a JAK1 gain-of-function mutant. Proliferation of MOHITO cells expressing both the activating JAK1 A634D mutant and either a CD45 targeting or a control shRNA was followed over a period of 72 hours and normalized to the control shRNA. The average ± SEM of 3 repeats is shown. *Statistical significance. (D) Overexpression of CD45 reduces sensitivity of HEK293T cells to IFN-α treatment. HEK293T cells transfected with empty vector or human CD45 (isoform R0) were stimulated with 500 U/mL IFN-α and harvested at different time points. STAT3 phosphorylation was assayed on Western blot and quantified relative to total STAT3.
Effects of CD45 knockdown and overexpression. (A) Knockdown of CD45 increases sensitivity of the human T-ALL cell line KE-37 to IL-7 stimulation. KE-37 cells were electroporated with a CD45 targeting or control siRNA. At 72 hours after electroporation, a fraction of the cells were stimulated with 10 ng/mL IL-7 for 10 minutes. Phosphorylation of STAT5 was assayed by Western blot (left) and quantified (right). (B) Knockdown of CD45 increases JAK/STAT signaling in MOHITO cells transformed by JAK1 or IL-7R gain-of-function mutants. MOHITO cells transformed by activating JAK1 or IL-7R mutants were transduced with a CD45 targeting or control shRNA. Phosphorylation of JAK1 and STAT5 was assayed by Western blot (left) and quantified (right). (C) Knockdown of CD45 increases proliferation of MOHITO cells transformed by a JAK1 gain-of-function mutant. Proliferation of MOHITO cells expressing both the activating JAK1 A634D mutant and either a CD45 targeting or a control shRNA was followed over a period of 72 hours and normalized to the control shRNA. The average ± SEM of 3 repeats is shown. *Statistical significance. (D) Overexpression of CD45 reduces sensitivity of HEK293T cells to IFN-α treatment. HEK293T cells transfected with empty vector or human CD45 (isoform R0) were stimulated with 500 U/mL IFN-α and harvested at different time points. STAT3 phosphorylation was assayed on Western blot and quantified relative to total STAT3.
In conclusion, we have identified and validated CD45 inactivating mutations, which demonstrate its function as a tumor suppressor gene in T-ALL. Notably, these loss-of-function mutations all occurred together with activating mutations in pathways that are negatively regulated by CD45 (supplemental Table 1): CD45 R751* with a JAK1 Y652H gain-of-function mutant in patient TLE-28,23 CD45 W764* with an activating IL-7R insertion in DND-41, and CD45 G863R with activating LCK mutations in HSB-2.5,24 Furthermore, our data indicate that loss of CD45 renders the JAK/STAT signaling pathway more susceptible to activation, either by cytokines or by oncogenic proteins, and can affect cancer cell proliferation. Although CD45-inactivating mutations appear to be quite rare, it has been reported that CD45 expression is extremely low or undetectable in 3.7% of pediatric T-ALL and 12.9% of pediatric B-cell precursor ALL patients,25 suggesting that additional mechanisms may exist for the inactivation of CD45.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the FWO-Vlaanderen (G.0287.07, J.C.), the Foundation against Cancer (SCIE2006-34, J.C.), the European Research Council (starting grant, J.C.), and KU Leuven (concerted action grant, J.C. and P.V.). Work in the Macintyre laboratory was supported by grants from the Association Laurette Fugain and the regional committee of the Ligue contre le Cancer. P.V. is a senior clinical investigator of FWO-Vlaanderen. M.T. is supported by the Associazione Italiana per la Ricerca sul Cancro (IG 2009-8803). M.P. is supported by the Agency for Innovation by Science and Technology in Flanders, Flanders, Belgium.
Authorship
Contribution: M.P. designed and performed research, analyzed data, and wrote the manuscript; M.K. and V.G. performed research, analyzed data, and wrote the manuscript; E.G. and K.D.K. analyzed data and wrote the manuscript; M.T., R.F., J.S., A.U., B.C., V.A., P.V., and E.M. contributed reagents and wrote the manuscript; and J.C. supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Cools, Center for Human Genetics, VIB11, Gasthuisberg O&N4, Herestraat 49 (Box 602), 3000 Leuven, Belgium; e-mail: jan.cools@cme.vib-kuleuven.be.