Abstract
The impact of a FLT3-internal tandem duplication (FLT3ITD) on prognosis of patients with acute myeloid leukemia (AML) is dependent on the ratio of mutated to wild-type allele. In 648 normal karyotype (NK) AML patients, we found a significant independent effect of the quantitative FLT3ITD mRNA level—measured as (FLT3ITD/wtFLT3)/(FLT3ITD/wtFLT3 + 1)—on outcome. Moreover, this effect was clearly seen in 329 patients with a mutated NPM1 gene (NPM1+), but not in 319 patients without a NPM1 mutation (wtNPM1). In a multivariate Cox regression model, the quantitative FLT3ITD mRNA level showed an independent prognostic impact on overall survival (OS) and relapse-free survival (RFS) only in the NPM1+ subgroup (OS: hazard ratio, 5.9; [95% confidence interval [CI]: 3.1-11.2]; RFS: hazard ratio, 7.5 [95% CI: 3.4-16.5]). The FLT3ITD mRNA level contributes to relapse risk stratification and might help to guide postremission therapy in NPM1-mutated AML.
Introduction
The prognosis of normal karyotype–acute myeloid leukemia (NK-AML) is influenced by the presence of gene mutations. NPM1 has been shown to be the most common single mutated gene in NK-AML occurring with a frequency of ∼ 50%. Combinations of NPM1 mutations with FLT3-internal tandem duplication (FLT3ITD) have been described in ∼ 20% of patients with NK-AML.1,2 The positive prognostic impact of the NPM1+ on outcome is mainly evident in patients lacking a FLT3ITD. Approximately 60% of patients carrying the NPM1+/FLT3–wild-type genotype survive > 10 years.3,4 The NPM1+ NK-AML has been classified as an own entity of favorable prognosis in the revised World Health Organization and European LeukemiaNet classifications.5,6 Since 2001, there have been reports that not only the presence of a FLT3ITD per se, but also the FLT3ITD/FLT3–wild-type (wtFLT3) ratio is essential for prognosis.7,8 The aim of our work was to assess the influence of the FTL3ITD mRNA level according to the mutation status of NPM1.
Methods
Patients
Our analyses were based on patients with NK-AML treated within the AML Cooperative Group 99 study.9 Patients were randomly assigned for induction therapy with either TAD (thioguanine, conventional-dose AraC, daunorubicin) followed by HAM (high-dose AraC, mitoxantrone) or 2 courses of HAM. As consolidation therapy in first complete remission (CR), allogeneic transplantation from an unrelated donor was recommended for high-risk patients < 60 years whereas all other patients received treatment with TAD and maintenance therapy.9
End points
Overall survival (OS) was calculated from randomization to death from any cause or to the latest follow-up. Relapse-free survival (RFS) was determined from the first day of CR until relapse or death in CR.
Molecular analyses
Mutation analyses of NPM1, FLT3ITD, FLT3–tyrosine kinase domain (FLT3TKD), MLL–partial tandem duplication (MLL-PTD), and CEBPA were performed according to standard protocols previously described.10-12 FLT3 mRNA RT-PCR and PCR were performed according to standard protocols.13 Labeled PCR products were electrophoresed on ABI 3100 (Applied Biosystems) according to protocol. The data were collected and analyzed with Genescan and Genotyper software (Applied Biosystems). The ratio of FLT3ITD mRNA to wtFLT3 mRNA was calculated as previously published.8,14 The amount of FLT3ITD mRNA in relation to the entire FLT3 transcript signal was defined as: quantitative “FLT3ITD mRNA level” = (FLT3ITD/wtFLT3)/(FLT3ITD/wtFLT3 + 1).
Statistical analyses
Univariate Cox regression for OS was first performed in the complete cohort to evaluate the prognostic value of the quantitative FLT3ITD mRNA level, independent of NPM1. For visualization of significant effects, we grouped patients according to the FLT3ITD mRNA level using 5 potential threshold values. To reduce the potential bias of data-derived cutpoints, we fixed the biologically meaningful thresholds 0.00, to distinguish between FLT3ITD and wtFLT3, 0.50, indicating a heterozygous mutation, and 1.00, indicating complete wild-type loss. In addition, we investigated the values 0.25 and 0.75 as potential thresholds. Very small patient groups (≤ 5%) were combined to the next larger adjacent group.
Multiple Cox regression using the quantitative FLT3ITD mRNA level, together with its interaction with NPM1 and clinical and molecular characteristics, was performed for OS and RFS. Kaplan-Meier estimation for OS and RFS and multiple Cox regression was also performed separately for NPM1+ and wtNPM1 patients. A significance level of 5% was used.
Results and discussion
Analyses were performed in 648 of 802 patients treated within the AMLCG99 trial (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Patients (119 of 648) received allogeneic transplantation in first CR. Median follow-up for OS was 62.3 months. Median OS was 20.4 months with 414 events. In 427 of 648 (66%) patients in CR, median RFS was 18.0 months. In 173 of 648 FLT3ITD-mutated patients, the median FLT3ITD level was 0.42 (0.02-1.00). Patient characteristics are summarized in supplemental Tables 1 and 2.
Impact of FLT3ITD mutation level on OS and definition of thresholds
Univariate Cox regression showed a significant impact of the FLT3ITD mRNA level on OS (hazard ratio of 1.12 for a FLT3ITD mutation level increased by 0.10, 95% confidence interval [CI], 1.08-1.17, P < .0001). Grouping patients using the prespecified threshold values, median OS for FLT3ITD mRNA level 0.00 (n = 471 of 648; 73%), 0.01-0.24 (n = 31 of 648; 5%), 0.25-0.49 (n = 91 of 648; 14%), 0.50-0.74 (n = 38 of 648; 6%), and 0.75-1.00 (n = 17 of 648; 3%) were 26, 24, 12, 8, and 8 months. The threshold level of 1.00 was excluded because only 7 patients had a complete wild-type loss. Because of the low patient number, FLT3ITD-positive patients with a level below 0.25 were combined with those with a level between 0.25 and 0.50 into a low-level (0.01-0.49) FLT3ITD group. Similarly, patients with a positive FLT3ITD mRNA level ≥ 0.50 were combined to one high-level (0.50-1.00) group. Finally, only the biologic meaningful cutpoints 0.00 and 0.50 were retained. Median OS in FLT3ITD-negative (73%), low-level (19%), and high-level FLT3ITD (8%) were 26.2, 15.6, and 7.8 months, respectively (P < .001).
Impact of FLT3ITD mutation level on outcome according to NPM1 mutation status
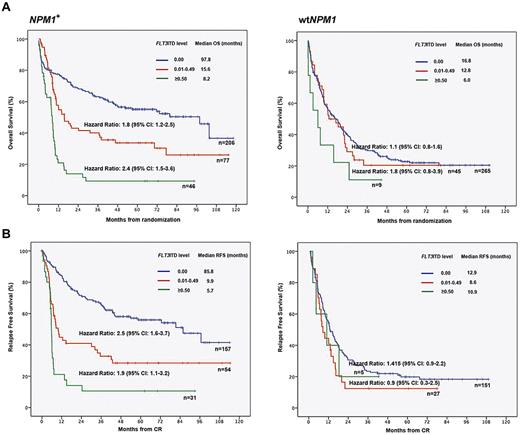
In the NPM1-mutated cohort, median OS was 97.8 months in the FLT3ITD-negative, 15.6 months in the low-level (0.01-0.49), and 8.2 months in the high-level FLT3ITD (0.50-1.00) group (P < .001, Figure 1A). Significant differences between these risk groups were evident regarding RFS (P < .001; Figure 1B). Median OS in wtNPM1 patients without a FLT3ITD, with a FLT3ITD level < 0.50 and ≥ 0.50 were not statistically different (16.8 months, 12.8 months, and 6.0 months, respectively, P = .133, Figure 1A). FLT3ITD mRNA level may not impact on survival in patients with wtNPM1, although this conclusion is limited by the low statistical power because of the relatively small number of patients with a high FLT3ITD mRNA level (n = 9; 1%).
Impact of FLT3ITD mutation level on outcome according to NPM1. (A) OS in patients with NPM1 mutation (N = 329) compared with NPM1 wild-type (n = 319). (B) RFS in patients with NPM1 mutation (N = 242) compared to NPM1 wild-type (n = 183). The significant impact of FLT3ITD mutation level on outcome was evident in NPM1-mutated AML. In NPM1-mutated AML, the effect of the FLT3ITD mRNA level displayed a dose-dependency. Thus, patients with a FLT3ITD level ≥ 0.50 showed the worst OS and RFS compared to patients with a FLT3ITD level between 0.01 and 0.49 and patients without a FLT3ITD. Differences between the score groups were highly significant (P ≤ .001).
Impact of FLT3ITD mutation level on outcome according to NPM1. (A) OS in patients with NPM1 mutation (N = 329) compared with NPM1 wild-type (n = 319). (B) RFS in patients with NPM1 mutation (N = 242) compared to NPM1 wild-type (n = 183). The significant impact of FLT3ITD mutation level on outcome was evident in NPM1-mutated AML. In NPM1-mutated AML, the effect of the FLT3ITD mRNA level displayed a dose-dependency. Thus, patients with a FLT3ITD level ≥ 0.50 showed the worst OS and RFS compared to patients with a FLT3ITD level between 0.01 and 0.49 and patients without a FLT3ITD. Differences between the score groups were highly significant (P ≤ .001).
In the multivariate Cox regression model with all 648 patients, the independent prognostic impact of the quantitative FLT3ITD mRNA level on outcome was detectable in NPM1+ patients (P < .001), but not in wtNPM1 (Table 1). This was true for both age subgroups (</≥ 60 years, data not shown). In multiple regression in NPM1+ patients, the FLT3ITD low-level group had an adjusted hazard ratio of 1.5 (95% CI, 0.96-2.3) for OS (P = .078), and the FLT3ITD high-level group an adjusted hazard ratio of 3.1 (95% CI 1.9-5.2, P < .001) compared with wtFLT3 (supplemental Table 3). Within wtNPM1 patients, the FLT3ITD mRNA level did not appear as an independent prognostic factor. Similar results were observed for RFS.
Multiple Cox regression models for OS and RFS
| Parameter . | . | Stratum . | OS, n = 508 . | RFS, n = 333 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||||
| Lower CL . | Upper CL . | Lower CL . | Upper CL . | |||||||
| NPM1 | pos vs neg | wtFLT3 | 0.3 | 0.2 | 0.4 | < .001 | 0.2 | 0.1 | 0.3 | < .001 |
| FLT3ITD mutation level | wtNPM1 | 1.1 | 0.4 | 3.0 | .789 | 0.6 | 0.2 | 2.2 | .436 | |
| FLT3ITD mutation level | NPM1+ | 5.9 | 3.1 | 11.2 | < .001 | 7.5 | 3.4 | 16.5 | < .001 | |
| Interaction NPM1*FLT3ITD mutation level | 5.2 | 1.7 | 15.3 | .003 | 12.7 | 3.0 | 55 | .001 | ||
| moCEBPA | vs wtCEBPA | 0.6 | 0.3 | 1.04 | .067 | 0.5 | 0.2 | 1.1 | .075 | |
| biCEBPA | vs wtCEBPA | 0.3 | 0.1 | 0.5 | < .001 | 0.3 | 0.1 | 0.6 | .001 | |
| FLT3TKD | pos vs neg | 1.4 | 0.9 | 2.3 | .149 | 1.0 | 0.5 | 2.2 | .890 | |
| MLL-PTD | pos vs neg | 0.9 | 0.6 | 1.4 | .699 | 0.8 | 0.4 | 1.4 | .377 | |
| WBC, ×106/L | 10-fold | 1.4 | 1.1 | 1.8 | .002 | 1.3 | 0.9 | 1.7 | .118 | |
| Platelets, ×106/L | 10-fold | 0.7 | 0.6 | 1.01 | .059 | 0.8 | 0.6 | 1.2 | .343 | |
| Hemoglobin level, mg/dL | +1 g/dL | 1.0 | 0.99 | 1.005 | .595 | 1.0 | 0.99 | 1.01 | .858 | |
| LDH, U/L | 10-fold | 1.2 | 0.8 | 1.8 | .503 | 1.2 | 0.7 | 2.1 | .616 | |
| BM blasts, % | +10% | 1.0 | 0.997 | 1.01 | .243 | 1.0 | 0.998 | 1.01 | .178 | |
| Age, y | +10 y | 1.4 | 1.2 | 1.5 | < .001 | 1.2 | 1.1 | 1.4 | < .001 | |
| Performance status, ECOG | 2-4 vs 0,1 | 1.3 | 0.996 | 1.6 | .054 | 1.2 | 0.9 | 1.6 | .310 | |
| Sex | Female vs male | 0.9 | 0.7 | 1.1 | .353 | 0.8 | 0.6 | 1.1 | .243 | |
| De novo AML | vs non–de novo | 0.9 | 0.7 | 1.3 | .709 | 0.9 | 0.6 | 1.4 | .781 | |
| Parameter . | . | Stratum . | OS, n = 508 . | RFS, n = 333 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||||
| Lower CL . | Upper CL . | Lower CL . | Upper CL . | |||||||
| NPM1 | pos vs neg | wtFLT3 | 0.3 | 0.2 | 0.4 | < .001 | 0.2 | 0.1 | 0.3 | < .001 |
| FLT3ITD mutation level | wtNPM1 | 1.1 | 0.4 | 3.0 | .789 | 0.6 | 0.2 | 2.2 | .436 | |
| FLT3ITD mutation level | NPM1+ | 5.9 | 3.1 | 11.2 | < .001 | 7.5 | 3.4 | 16.5 | < .001 | |
| Interaction NPM1*FLT3ITD mutation level | 5.2 | 1.7 | 15.3 | .003 | 12.7 | 3.0 | 55 | .001 | ||
| moCEBPA | vs wtCEBPA | 0.6 | 0.3 | 1.04 | .067 | 0.5 | 0.2 | 1.1 | .075 | |
| biCEBPA | vs wtCEBPA | 0.3 | 0.1 | 0.5 | < .001 | 0.3 | 0.1 | 0.6 | .001 | |
| FLT3TKD | pos vs neg | 1.4 | 0.9 | 2.3 | .149 | 1.0 | 0.5 | 2.2 | .890 | |
| MLL-PTD | pos vs neg | 0.9 | 0.6 | 1.4 | .699 | 0.8 | 0.4 | 1.4 | .377 | |
| WBC, ×106/L | 10-fold | 1.4 | 1.1 | 1.8 | .002 | 1.3 | 0.9 | 1.7 | .118 | |
| Platelets, ×106/L | 10-fold | 0.7 | 0.6 | 1.01 | .059 | 0.8 | 0.6 | 1.2 | .343 | |
| Hemoglobin level, mg/dL | +1 g/dL | 1.0 | 0.99 | 1.005 | .595 | 1.0 | 0.99 | 1.01 | .858 | |
| LDH, U/L | 10-fold | 1.2 | 0.8 | 1.8 | .503 | 1.2 | 0.7 | 2.1 | .616 | |
| BM blasts, % | +10% | 1.0 | 0.997 | 1.01 | .243 | 1.0 | 0.998 | 1.01 | .178 | |
| Age, y | +10 y | 1.4 | 1.2 | 1.5 | < .001 | 1.2 | 1.1 | 1.4 | < .001 | |
| Performance status, ECOG | 2-4 vs 0,1 | 1.3 | 0.996 | 1.6 | .054 | 1.2 | 0.9 | 1.6 | .310 | |
| Sex | Female vs male | 0.9 | 0.7 | 1.1 | .353 | 0.8 | 0.6 | 1.1 | .243 | |
| De novo AML | vs non–de novo | 0.9 | 0.7 | 1.3 | .709 | 0.9 | 0.6 | 1.4 | .781 | |
The independent prognostic impact of the FLT3ITD mutation level on OS and RFS was evaluated using multivariate Cox regression models. The FLT3ITD mutation level was introduced as a continuous parameter into the model. Due to the known interaction between NPM1 and FLT3ITD, an interaction term NPM1*FLT3ITD mutation level was included in the model. Besides the FLT3ITD mutation level, mutations of the molecular markers NPM1 (NPM1+), CEBPA (moCEBPA; biCEBPA), FLT3TKD, MLL-PTD, and the clinical parameters age, sex, ECOG performance status, AML de novo, WBC, platelet count, hemoglobin level, LDH, and amount of BM blasts were introduced into the model. The multivariate prognostic factors were identified using a logistic regression model with a significance level of 5%.
OS indicates overall survival; RFS, relapse-free survival; moCEBPA, monoallelic CEBPA mutation; biCEBPA, biallelic CEBPA mutation; TKD, tyrosine kinase domain; PTD, partial tandem duplication; ITD, internal tandem duplication; WBC, white blood count; LDH, lactate dehydrogenase; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; CI, confidence interval; CL, confidence limit; pos, positive; neg, negative; and AML, acute myeloid leukemia.
Whitman et al were the first to show that a complete loss of wtFLT3 was associated with worse outcome compared to patients without a FLT3ITD (wtFLT3/wtFLT3) or a heterozygous FLT3ITD (wtFLT3/FLT3ITD) mutation.7 Thiede et al defined the FLT3ITD/wt ratio as the relative proportion of the area under the curve (AUC) of mutant and wtFLT3 alleles (AUC-FLT3ITD/AUC-wtFLT3) in Genescan analysis. A FLT3ITD/wtFLT3 ratio above the median of the cohort was associated with an unfavorable prognosis.8 Median-defined risk groups have to be determined in large patient cohorts before a definite statement about individual prognosis can be made. In contrast, we defined the FLT3ITD mRNA level as the relative amount of FLT3ITD mRNA to the total FLT3 transcript, with a range from 0 (absence of mutation) to 1 (complete loss of wild type), facilitating the estimation of the FLT3ITD mutational load. This has the advantage of direct estimation of individual prognosis according to a patient's FLT3ITD mutant level and better comparability in different clinical studies.
The focus of our analyses was the investigation of the impact of the FLT3ITD mRNA level according to the NPM1 mutation status in NK-AML. Univariate and multivariate analyses demonstrated a distinct dose-dependent effect of the FLT3ITD mutant level on OS and RFS only in NPM1+, but not in wtNPM1 patients. In NPM1-mutated patients, multivariate analyses revealed a FLT3ITD level of 0.50 as cutoff between an intermediate group (26% long-term survivors) and a poor-risk group with 9% survivors in 7 years. In accordance with Whitman et al, these observations suggest different pathophysiologies of heterozygous FLT3ITD versus FLT3ITD with a complete loss of the wild-type allele.7
Our data suggest a significantly worse outcome with regard to OS and RFS for patients harboring an NPM1 mutation and higher FLT3ITD mRNA expression compared to those NPM1-mutated patients with a low FLT3ITD mRNA expression. Thus, the FLT3ITD mRNA level might guide the decision for allogeneic transplantation in NPM1+ AML. However, such a strategy should be prospectively evaluated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.S. performed statistical analysis and wrote the manuscript; E.H., M.U., A.H., and M.C.S. provided statistical support; S.S., A.D., T. Benthaus, G.M., E.Z., and P.M.K. performed molecular diagnostics; S.K.B., M.F.-B., C.B., J.B., and K.S. performed central diagnostics; W.E.B., T. Büchner, B.J.W., and W.H. were principal investigators of AMLCG99 study; and K.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karsten Spiekermann, MD, Department of Medicine III, University Hospital Grosshadern, Marchioninistr 15, 81377 Munich, Germany; e-mail: karsten.spiekermann@med.uni-muenchen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal