SF3B1 mutations were recently reported in myelodysplastic syndromes (MDSs), especially in the presence of ring sideroblasts (RSs). We sought to define the interaction between SF3B1 mutations, morphology, karyotype, and prognosis in MDS with more than or equal to 15% RS (MDS-RS). We studied 107 patients with MDS-RS, including 48 with refractory anemia with RS (RARS), 43 with refractory cytopenia with multilineage dysplasia (RCMD)-RS, 11 with refractory anemia with excess blasts-1 (RAEB1)–RS, and 5 with RAEB2-RS. SF3B1 mutations were detected in 53 (∼ 50%) patients: 35 RARS (73%), 16 RCMD-RS (37%), and 2 RAEB1-RS (18%). In univariate analysis, the presence of SF3B1 mutations was associated with better overall (P < .01) and leukemia-free (P < .01) survival; however, in both instances, significance was completely accounted for by World Health Organization morphologic risk categorization. In other words, when RARS and RCMD-RS were analyzed separately, there was no additional prognostic value from the presence or absence of SF3B1 mutations.
Introduction
The presence of more than or equal to 15% ring sideroblasts (RSs) constitutes the operational diagnosis of myelodysplastic syndromes with RS (MDS-RS), which includes refractory anemia with ring sideroblasts (RARSs).1,2 RSs are also seen in both acquired non-neoplastic conditions (eg, excess alcohol use, lead toxicity, copper or pyridoxine deficiency, isoniazid therapy) and hereditary sideroblastic anemias associated with a number of germline mutations involving δ-aminolevulinate synthase 2 (ALAS2),3 solute carrier family 25 member 38 (SLC25A38),4 glutaredoxin-5 (GLRX5),5 and the ATP-binding cassette subfamily B member 7 (ABCB7 gene, which encodes for a protein thought to enable mitochondrial iron transport)6 genes. The pathogenetic contribution of these genes in MDS-RS is not clear, although up-regulation of ALAS2 and down-regulation of ABCB7 have been reported in RARS.7,8 Recently, 3 groups have demonstrated recurrent somatic mutations of the splicing factor 3B subunit 1 (SF3B1) gene in a high proportion of patients with RARS (64%-68%-83%) or refractory cytopenia with multilineage dysplasia (RCMD)–RS (57%-76%).9,–11 SF3B1 is located on chromosome 2q33.1 and encodes for SF3B1, which is part of a multiprotein complex (SF3B associates with SF3A and 12S RNA to form the U2 small nuclear ribonucleoprotein particles) that is thought to play a role in pre-mRNA splicing and associated transcription.12 In the current study, we sought to accurately define the prevalence of SF3B1 mutations in different morphologic subcategories of MDS-RS and examine their phenotypic and prognostic correlates.
Methods
The current study was approved by the Mayo Clinic Institutional Review Board. Mayo Clinic databases and cell banks were queried to identify patients with MDS and ≥ 15% RS (MDS-RS). All study patients were required to have undergone bone marrow examination and cytogenetic evaluation at diagnosis. Pathology slides, including iron stains, were centrally rereviewed (C.A.H. and J.M.H.) to accurately quantify bone marrow RS percentage and confirm World Health Organization morphologic diagnosis and classification. Patients were risk-stratified according to the revised International Prognostic Scoring System (IPSS-R).13 All patients were also screened for JAK2, MPL, and IDH mutations, using previously described methods.14,–16
SF3B1 coding exon sequencing was performed using bone marrow- or granulocyte-derived DNA extract (QIAGEN), obtained at time of diagnosis, that was amplified using the primer sets detailed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Individual mutations were examined and excluded for occurrence in normal genetic variation by interrogation of the National Center for Biotechnology Information single nucleotide polymorphism database.17 All products were confirmed by 1% agarose gel, purified using QIAquick Spin Kit (QIAGEN), and submitted for sequencing using 2 ABI PRISM 3730xl DNA Analyzers (96-capillary). For SF3B1 exons 13 to 15 sequencing, 50 to 100 ng DNA was added to a 50-μL final volume mixture of: 5 μL 10× PCR Buffer (Roche Diagnostics), 1.5 μL of dNTP (10mM of each), 0.5 μL of Taq Polymerase (Roche Diagnostics), and 2 μL each of a 10μM stock of forward (SH3B1 FP 5′-TAGAGTGGAAGGCCGAGAGA-3′) and reverse (SH3B1 RP 5′- TTCAAGAAAGCAGCCAAACC-3′) primers, supplemented with PCR grade water. Parameters for this PCR amplification included an initial denaturation step of 95°C for 2 minutes, followed by 35 cycles of an initial melt at 95°C for 15 seconds, annealing at 60°C for 45 seconds, extension at 72°C for 70 seconds, and a final extension step of 72°C for 3 minutes. The products were purified with QIAquick Spin Kit (QIAGEN). Sequencing was performed bidirectionally. Standard statistical methods were used for parameter comparison and survival curves were prepared by the Kaplan-Meier method and compared by the log-rank test. Cox proportional hazard regression model was used for multivariable analysis. P values < .05 were considered significant. The Stat View (SAS Institute) statistical package was used for all computations.
Results and discussion
The study included 107 patients with MDS-RS: 48 RARS, 43 refractory cytopenia with multilineage dysplasia and more than or equal to 15% RS (RCMD-RS), 11 refractory anemia with excess blasts (RAEB)-1 and more than or equal to 15% RS (RAEB1-RS), and 5 with RAEB2-RS; 7, 5, and zero patients displayed IDH (IDH1-1, IDH2-6), JAK2, or MPL mutations. Table 1 outlines the clinical and laboratory features and subsequent events in all 107 study patients with MDS-RS, stratified by the presence or absence of SF3B1 mutations.
We initially sequenced all SF3B1 coding exons in 10 RARS cases and identified 7 patients with mutations that clustered in exons 14 and 15. We subsequently sequenced all SF3B1 exons in an additional 5 patients with RCMD-RS who were negative for SF3B1 mutations in exons 14 and 15 and found no additional mutations outside of exons 14 and 15. Accordingly, only exons 13 to 15 were sequenced in the remaining patients.
SF3B1 mutations (all heterozygous) were detected in 53 (49.5%) patients: 34 (64%) K700E, 6 (11%) K666N/Q/R,4 E622D, 3 H662D/Q, 2 Y623C, 2 R625C/L, and 2 T663I. One patient each displayed both SF3B1K700E and IDH2R140Q or SF3B1K700E and JAK2V617F. SF3B1 mutational frequencies were 73% for RARS, 37% for RCMD-RS, 18% for RAEB1-RS, and 0% for RAEB2-RS (P < .01). SF3B1 mutations clustered with normal karyotype (P < .01), and only 7 (13%) of the 53 SF3B1 mutations were accompanied by an abnormal karyotype other than −Y, and 3 of the 7 associated abnormalities involved del(20q) (supplemental Table 2). SF3B1 mutations correlated with lower IPSS-R risk category (P < .01) and a higher platelet count (P < .01; Table 1).
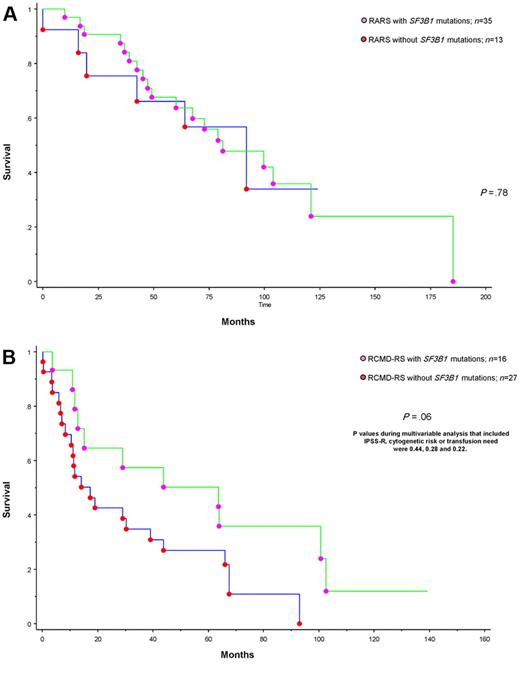
To date, 75 (∼ 70%) deaths and 11 (∼ 10%) leukemic transformations have been recorded. Median follow-up for living patients was 82 months. In univariate analysis, survival was predicted by advanced age (P = .04), IPSS-R (P < .01), World Health Organization morphologic category (P < .01), karyotype (P < .01), and transfusion need (P < .01). In multivariable analysis, only age, IPSS-R, and transfusion need remained significant (supplemental Table 3). In addition, in univariate analysis, the presence of SF3B1 mutations was significantly associated with better overall (P < .01) and leukemia-free survival (P < .01); however, in both instances, significance was completely accounted for by World Health Organization morphologic risk categorization, and morphology-adjusted P values were .6 and .2, respectively (supplemental Tables 3 and 4). In other words, when RARS and RCMD were analyzed separately, there was no additional prognostic value from the presence or absence of SF3B1 mutations (Figure 1; P = .78 and P = .06). The latter borderline significance, in the context of RCMD-RS, was completely lost during multivariable analysis that included, as a covariate, IPSS-R, cytogenetic risk stratification, or transfusion need; the respective P values were .44, .28, and .22.
Survival data for patients with MDS and ring sideroblasts stratified by the presence or absence of SF3B1 mutation. (A) Survival data for 48 patients with RARS stratified by the presence or absence of SF3B1 mutations. (B) Survival data for 43 patients with RCMD-RS stratified by the presence or absence of SF3B1 mutations.
Survival data for patients with MDS and ring sideroblasts stratified by the presence or absence of SF3B1 mutation. (A) Survival data for 48 patients with RARS stratified by the presence or absence of SF3B1 mutations. (B) Survival data for 43 patients with RCMD-RS stratified by the presence or absence of SF3B1 mutations.
The current study confirms the recent discovery demonstrating the high prevalence of SF3B1 mutations in RARS and RCMD-RS.9,–11 In addition, we show that the apparent association of SF3B1 mutations with good prognosis was the function of its significant alignment with RARS and that the mutation by itself did not carry additional prognostic relevance. Yoshida et al reported the occurrence of heterozygous SF3B1 mutations in 19 (82.6%) of 23 patients with RARS and 38 (76%) of 50 patients with RCMD-RS.11 Mutations involving the SRSF2 (5.5%) and ZRSR2 (1.4%) genes were also seen in the Yoshida et al study.11 Papaemmanuil et al demonstrated heterozygous SF3B1 mutations in 40 (68%) of 59 patients with RARS and in 13 (57%) of 23 patients with RCMD-RS.9 We suspect differences in interpretation of morphology and disease designation as RARS versus RCMD accounted for the discrepancy in SF3B1 mutational frequency among these studies. In the latter study, SF3B1-mutated patients displayed higher white cell, and platelet counts and appeared to have a more favorable prognosis.9 A number of scientific and practical issues await clarification regarding SF3B1 mutations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.M.P. and A.T. designed the study, contributed patients, collected data, performed the statistical analysis, and wrote the paper; T.L.L. participated in study design, primer design, and sequence analysis; J.M.H. and C.A.H. reviewed histopathology; R.A.K. and R.P.K. reviewed cytogenetic information; G.G.-M. participated in study design and data analysis; D.P.S. collected data and participated in study design and data analysis; A.P. collected data and participated in study design, primer design, and sequence analysis; and all authors approved the final draft of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ayalew Tefferi, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: tefferi.ayalew@mayo.edu.