Abstract
DNMT3A mutations are associated with poor prognosis in acute myeloid leukemia (AML), but the stability of this mutation during the clinical course remains unclear. In the present study of 500 patients with de novo AML, DNMT3A mutations were identified in 14% of total patients and in 22.9% of AML patients with normal karyotype. DNMT3A mutations were positively associated with older age, higher WBC and platelet counts, intermediate-risk and normal cytogenetics, FLT3 internal tandem duplication, and NPM1, PTPN11, and IDH2 mutations, but were negatively associated with CEBPA mutations. Multivariate analysis demonstrated that the DNMT3A mutation was an independent poor prognostic factor for overall survival and relapse-free survival in total patients and also in normokaryotype group. A scoring system incorporating the DNMT3A mutation and 8 other prognostic factors, including age, WBC count, cytogenetics, and gene mutations, into survival analysis was very useful in stratifying AML patients into different prognostic groups (P < .001). Sequential study of 138 patients during the clinical course showed that DNMT3A mutations were stable during AML evolution. In conclusion, DNMT3A mutations are associated with distinct clinical and biologic features and poor prognosis in de novo AML patients. Furthermore, the DNMT3A mutation may be a potential biomarker for monitoring of minimal residual disease.
Introduction
DNMT3A encodes the enzyme DNA methyltransferase (DNMT) 3A, which catalyzes the addition of methyl groups to the cytosine residue of CpG dinucleotides in DNA.1,2 DNMT3A contains 3 main structure domains: an ATRX, DNMT3, and DNMT3L–type zinc finger domain, a proline-tryptophan-tryptophan-proline domain, and the methyltransferase (MTase) domain.1 The proline-tryptophan-tryptophan-proline domain is responsible for targeting the enzyme to nucleic acid, whereas the zinc finger domain mediates protein-protein interactions with the transcription factors Myc and RP58, the heterochromatin protein HP1, histone deacetylases, and the histone methyltransferase Suv39h1.2 Recently, mutations in DNMT3A were identified in patients with AML, myelodysplastic syndromes, and myeloproliferative neoplasms.3–7 The incidences of this mutation in AML varied: 4.1% in a Japanese study,8 9% (among all AML, including M4/M5 and other subtypes) in a Chinese study,9 and approximately 20% in 2 Western studies.3,4 Whether there is a geographic difference in the incidence of DNMT3A mutations needs to be determined. Furthermore, sequential analyses to evaluate the stability of DNMT3A mutations during the clinical course were limited to a small number of patients. In the present study, we investigated the DNMT3A mutation in 506 patients with de novo AML and analyzed its interactions with 16 other gene alterations. Sequential analysis of the DNMT3A mutation during the clinical course was also performed on 138 patients to investigate the stability and pathogenic role of this mutation in AML. Further, to better stratify AML patients into different risk groups, a scoring system integrating DNMT3A mutations with 8 other prognostic factors, including age, WBC count, cytogenetics, NPM1/FLT3 internal tandem duplication (NPM1/FLT3-ITD), CEBPA, AML1/RUNX1, WT1, and IDH2 mutations, into survival analysis was proposed.
Methods
Subjects
This study was approved by the institutional review board of the National Taiwan University Hospital (NTUH), and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. From March 1995 to December 2008, a total of 506 adult patients who were newly diagnosed as having de novo AML at NTUH and had enough cryopreserved cells for analysis were enrolled consecutively. Patients with antecedent hematologic diseases or therapy-related AML were excluded. Diagnosis and classification of AML were made according to the French-American-British (FAB) Cooperative Group Criteria.
In total, 363 (71.7%) patients received standard induction chemotherapy (idarubicin 12 mg/m2/d on days 1-3 and cytarabine 100 mg/m2/d on days 1-7) and then consolidation chemotherapy with 2-4 courses of high-dose cytarabine (2000 mg/m2 every 12 hours on days 1-4 for a total of 8 doses), with or without an anthracycline (idarubicin or Novantrone), after achieving complete remission (CR).10,11 The patients with acute promyelocytic leukemia (M3 subtype) received concurrent all-trans retinoic acid and chemotherapy. The remaining 143 patients received palliative therapy with supportive care and/or low-dose chemotherapy because of underlying comorbidities or based on patient decision. Forty-five patients received allogeneic hematopoietic stem cell transplantation (HSCT) in first CR.
Cytogenetics
BM cells were harvested directly or after 1-3 days of unstimulated culture, as described previously.12 Metaphase chromosomes were banded with the trypsin-Giemsa technique and karyotyped according to the International System for Human Cytogenetic Nomenclature.
Immunophenotype analysis
A panel of mAbs to myeloid-associated antigens, including CD13, CD33, CD11b, CD15, CD14, and CD41a, as well as lymphoid-associated antigens, including CD2, CD5, CD7, CD19, CD10, and CD20, and lineage-nonspecific antigens HLA-DR, CD34, and CD56, were used to characterize the phenotypes of the leukemia cells, as described previously.13
Mutation analysis
Mutation analysis of DNMT3A exons 2-23 was performed by PCR and direct sequencing as described previously.4 Abnormal sequencing results were confirmed by at least 2 repeated analyses. Sequential analysis of the DNMT3A mutation during the clinical course was performed in 316 samples from 138 patients. Mutation analyses of 16 other relevant molecular marker genes, including class I mutations, such as FLT3-ITD and FLT3-TKD,13 N-RAS,14 K-RAS,14 JAK2,14 KIT,15 and PTPN1116 mutations, and class II mutations, such as MLL-PTD,17 CEBPA,18 and AML1/RUNX111 mutations, as well as NPM1,19 WT1,10 ASXL1,20 IDH1,21 IDH222 (including R140 and R172 mutations), and TET223 mutations, were performed as described previously. To detect DNMT3A mutation at diagnosis, we used DNA amplified in vitro from BM cells with the Illustra GenomiPhi V2 DNA-amplification kit as described by the manufacturer (GE Healthcare). All mutations detected were verified in the original nonamplified samples. All nucleotide alterations causing premature truncation of the DNMT3A proteins (nonsense or frame-shift mutations) were regarded as true mutations. Missense mutations were regarded as true only if they were documented in other studies or could be verified by sequencing of normal somatic tissues or matched remission BM samples.
TA cloning analysis
For patients with double mutations, Taq polymerase-amplified (TA) cloning was performed to determine whether the 2 mutations were in the same or different alleles, as described previously.15 Briefly, the cDNA was amplified to cover both mutations and the PCR products were then cloned into the TA-cloning vector pGEM-T Easy (Promega) and more than 10 clones were selected for sequencing.
Statistical analysis
The discrete variables of patients with and without gene mutation were compared using the χ2 tests, but if the expected values of contingency tables were smaller than 5, the Fisher exact test was used. If the continuous data were not normally distributed, Mann-Whitney U tests were used to compare continuous variables and medians of distributions. To evaluate the impact of the DNMT3 mutation on clinical outcome, only the patients who received conventional standard chemotherapy were included in the analysis.10,11 Overall survival (OS) was measured from the date of first diagnosis to the date of last follow-up or death from any cause, whereas relapse was defined as a reappearance of at least 5% leukemic blasts in a BM aspirate or new extramedullary leukemia in patients with a previously documented CR.24 Relapse-free survival (RFS) was measured from the date of attaining the leukemia-free state until the date of AML relapse or death from any cause, whichever occurred first. Cox regression survival estimation was used to plot survival curves and to test the differences between groups. Multivariate Cox proportional hazard regression analysis was used to investigate independent prognostic factors for OS and RFS. The proportional hazards assumption (constant hazards assumption) was examined using time-dependent covariate Cox regression before conducting multivariate Cox proportional hazard regression. The variables including age, WBC counts, karyotype, NPM1/FLT3-ITD, WT1, CEBPA, AML1/RUNX1, TET2, ASXL1, IDH2, and DNMT3A mutations were used as covariates. Those patients who received HSCT were censored at the time of transplantation in survival analysis to ameliorate the influence of the treatment.10,11 P < .05 was considered statistically significant. All statistical analyses were performed with SPSS Version 18 software and Statsdirect (2.7.8b, 2011).
Results
DNMT3A mutations in patients with de novo AML
Excluding the 8 single-nucleotide polymorphisms (P9P, S267S, G291G, A398A, P385P, L422L, V435V, and V563V) that were detected in 316 patients but did not alter the amino acid residues, and the 7 missense mutations (C586W, P896L, G543C, Y735C, A644T, G699D, and G707D) that were found in 6 patients but had uncertain biologic significance (because they were not reported previously and could not be verified because of lack of matched BM samples at CR), DNMT3A mutations at 30 different positions were identified in 70 patients (Table 1 and Figure 1). Twelve were missense mutations, 8 were nonsense mutations, 9 were frame-shift mutations, and 1 was an in-frame mutation. The most common mutation was R882H (n = 26), followed by R882C (n = 15), R882S (n = 3), R736H (n = 3), and R320X (n = 2). All other mutations were detected in only 1 patient each. Mutations at exon 23 occurred in 47 patients, including the 44 patients with R882 mutations. Four patients had double heterozygous mutations (patients 43, 64, 65, and 68); in 1 of them (patient 64), the 2 mutations were confirmed to be biallelic by DNA PCR and TA cloning, and for the other 3, the nature of the double mutations was not verified by this method because the 2 mutations were located in different exons too far apart to be amplified by a single-DNA PCR reaction. The remaining 66 patients showed only one mutation; all were heterozygous.
Mutation patterns in 70 patients with DNMT3A mutations at diagnosis
| UPN . | Age, y/Sex . | FAB . | Karyotype . | Location . | DNMT3 mutation . | Other accompanying gene mutations . | |
|---|---|---|---|---|---|---|---|
| DNA change . | Protein change . | ||||||
| 1 | 79/M | M5 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, MLL/PTD, IDH2 |
| 2 | 77/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | AML1/RUNX1 |
| 3 | 64/M | M5 | NM | 23 | c.2646G > A | p.R882H | PTPN11, NPM1 |
| 4 | 73/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | IDH2 |
| 5 | 16/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/TKD, NPM1 |
| 6 | 41/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 7 | 80/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | PTPN11, NPM1 |
| 8 | 61/F | M5 | 46,XX t(5;17)(q33;q21) | 23 | c.2645C > T | p.R882C | FLT3/TKD, NPM1 |
| 9 | 46/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 10 | 35/F | M1 | 46,XX | 18 | c.2120delG | p.G707AfsX72 | NRAS, IDH1 |
| 11 | 82/M | M0 | ND | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD, AML1/RUNX1 |
| 12 | 79/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, FLT3/TKD, MLL/PTD, TET2 |
| 13 | 51/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/TKD, NPM1 |
| 14 | 55/M | M4 | 46,XY | 16 | c.1865_1866 insGT | p.Y623FfsX29 | NPM1 |
| 15 | 54/M | M4 | 46,XY | 8 | c.890G > A | p.W297X | PTPN11, ASXL1 |
| 16 | 68/M | M2 | 46,XY | 23 | c.2645C > A | p.R882S | FLT3/ITD, NPM1 |
| 17 | 45/F | M5 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/TKD, AML1/RUNX1, IDH2 |
| 18 | 54/F | M2 | 46,XX | 23 | c.2646G > A | p.R882H | NRAS, NPM1 |
| 19 | 87/M | M4 | 46,XY | 23 | c.2606delG | p.G869VfsX12 | FLT3/TKD, NPM1 |
| 20 | 51/F | M4 | 47,XX,+i(11)(q10) | 20 | c.2389A > T | p.N797Y‡ | ASXL1, IDH2 |
| 21 | 78/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 22 | 38/F | M5 | 46,XX | 23 | c.2645C > T | p.R882C | NRAS, NPM1, IDH1 |
| 23 | 72/F | M2 | 46,XX,del(20)(q11q13) | 13 | c.1477delA | p.I493SfsX158 | FLT3/ITD, NPM1 |
| 24 | 65/F | M5 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD |
| 25 | 42/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, AML1/RUNX1, ASXL1 |
| 26 | 78/M | M2 | 46,X,−Y,+4 | 19 | c.2246_2249del | p.R749PfsX29 | FLT3/TKD, NPM1 |
| 27 | 75/F | M1 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 28 | 51/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 29 | 60/F | M1 | 46,XX,t(9;22)(q34;q11) | 18 | c.2113A > T | p.I705F§ | IDH1 |
| 30 | 73/F | M1 | 46,XX | 23 | c.2646G > A | p.R882H | CEBPA, TET2 |
| 31 | 22/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 32 | 38/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 33 | 31/F | M5 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 34 | 46/M | M4 | 45X,−Y | 23 | c.2645C > A | p.R882S | NRAS, FLT3/ITD, NPM1 |
| 35 | 80/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD |
| 36 | 52/M | M1 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, IDH2 |
| 37 | 44/M | M2 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 38 | 33/F | M1 | 46,XX | 8 | c.958C > T | p.R320X | FLT3/TKD, NPM1 |
| 39 | 42/M | M4 | 45,X,−Y | 15 | c.1816C > T | p.Q606X | NPM1 |
| 40 | 78/F | M2 | 46,XX | 19 | c.2255_2257del | p.F752del | FLT3/ITD, NPM1 |
| 41 | 75/F | M2 | 47,XX,del(5)(q22q35),+8 | 8 | c.958C > T | p.R320X | IDH2 |
| 42 | 49/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 43 | 78/M | M4 | 46,XY | 4 | c.315C > A | p.S105R | PTPN11 |
| 44 | 64/M | M5 | 46,XY | 23 | c.2645C > A | p.R882S | PTPN11, MLL/PTD |
| 45 | 40/M | M5 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 46 | 75/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | NRAS, NPM1, TET2 |
| 47 | 58/F | M2 | 46,XX | 23 | c.2645C > T | p.R882C | NPM1 |
| 48 | 48/F | M1 | 47,XX,+8 | 8 | c.1001delG | p.G334AfsX11 | CEBPA, IDH2 |
| 49 | 67/M | M5 | 47,XY,+8 | 23 | c.2645C > T | p.R882C | PTPN11, KRAS, AML1/RUNX1, IDH2 |
| 50 | 51/M | M2 | 46XY | 23 | c.2646G > A | p.R882H | IDH2 |
| 51 | 85/M | M5 | 46,XY | 7 | c.767_770del | P256LfsX59 | FLT3/ITD, NPM1, WT1 |
| 52 | 85/M | M1 | 45,XY,−7 | 4 | c.327_328insG | Q110AfsX14 | |
| 53 | 67/M | M8 | 47,XY,+8 | 23 | c.2646G > A | p.R882H | NRAS, IDH2 |
| 54 | 35/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 55 | 47/F | M2 | 46,XX | 23 | c.2646G > A | p.R882H | ASXL1, IDH2 |
| 56 | 50/F | M1 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD |
| 57 | 86/M | M5 | 46,XY | 8 | c.866delG | p.G289AfsX26 | FLT3/ITD |
| 58 | 69/M | M1 | NM | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1, CEBPA |
| 59 | 75/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | AML1/RUNX1 |
| 60 | 79/F | M4 | Cplx* | 22 | c.2510C > G | p.S837X | |
| 61 | 61/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | NPM1, WT1, TET2 |
| 62 | 37/F | M2 | 46,XX | 19 | c.2312G > A | p.R771Q‡ | NPM1, TET2 |
| 63 | 70/M | M2 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1, IDH2 |
| 64 | 46/M | M4 | 46,XY | 19 | c.2182G > C, c.2191T > C | p.G728R‡, p.F731L‡ | FLT3/ITD |
| 65 | 69/M | M4 | 47,XY,+X | 8 | c.941G > A | p.W314X | NRAS, FLT3/TKD, AML1/RUNX1, IDH2 |
| 19 | c.2207G > A | p.R736H† | |||||
| 66 | 38/F | M2 | 46,XX | 19 | c.2207G > A | p.R736H† | FLT3/ITD, NPM1, IDH1 |
| 67 | 66 | M1 | 47,XY,del(5)(q31q35), der(7)t(5;7)(q13;q11),+8 | 15 | c.1792C > T | p.R598X | IDH2 |
| 68 | 81 | M4 | 46,XY | 17 | c.2032C > T | p.Q678X | NRAS, TET2 |
| 19 | c.2210T > A | p.L737H‡ | |||||
| 69 | 50 | M4 | 46,XX | 15 | c.1903C > T | p.R635W† | PTPN11, NPM1, IDH2 |
| 70 | 84 | M0 | ND | 19 | c.2207G > A | p.R736H† | AML1/RUNX1, IDH2 |
| UPN . | Age, y/Sex . | FAB . | Karyotype . | Location . | DNMT3 mutation . | Other accompanying gene mutations . | |
|---|---|---|---|---|---|---|---|
| DNA change . | Protein change . | ||||||
| 1 | 79/M | M5 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, MLL/PTD, IDH2 |
| 2 | 77/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | AML1/RUNX1 |
| 3 | 64/M | M5 | NM | 23 | c.2646G > A | p.R882H | PTPN11, NPM1 |
| 4 | 73/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | IDH2 |
| 5 | 16/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/TKD, NPM1 |
| 6 | 41/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 7 | 80/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | PTPN11, NPM1 |
| 8 | 61/F | M5 | 46,XX t(5;17)(q33;q21) | 23 | c.2645C > T | p.R882C | FLT3/TKD, NPM1 |
| 9 | 46/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 10 | 35/F | M1 | 46,XX | 18 | c.2120delG | p.G707AfsX72 | NRAS, IDH1 |
| 11 | 82/M | M0 | ND | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD, AML1/RUNX1 |
| 12 | 79/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, FLT3/TKD, MLL/PTD, TET2 |
| 13 | 51/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/TKD, NPM1 |
| 14 | 55/M | M4 | 46,XY | 16 | c.1865_1866 insGT | p.Y623FfsX29 | NPM1 |
| 15 | 54/M | M4 | 46,XY | 8 | c.890G > A | p.W297X | PTPN11, ASXL1 |
| 16 | 68/M | M2 | 46,XY | 23 | c.2645C > A | p.R882S | FLT3/ITD, NPM1 |
| 17 | 45/F | M5 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/TKD, AML1/RUNX1, IDH2 |
| 18 | 54/F | M2 | 46,XX | 23 | c.2646G > A | p.R882H | NRAS, NPM1 |
| 19 | 87/M | M4 | 46,XY | 23 | c.2606delG | p.G869VfsX12 | FLT3/TKD, NPM1 |
| 20 | 51/F | M4 | 47,XX,+i(11)(q10) | 20 | c.2389A > T | p.N797Y‡ | ASXL1, IDH2 |
| 21 | 78/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 22 | 38/F | M5 | 46,XX | 23 | c.2645C > T | p.R882C | NRAS, NPM1, IDH1 |
| 23 | 72/F | M2 | 46,XX,del(20)(q11q13) | 13 | c.1477delA | p.I493SfsX158 | FLT3/ITD, NPM1 |
| 24 | 65/F | M5 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD |
| 25 | 42/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, AML1/RUNX1, ASXL1 |
| 26 | 78/M | M2 | 46,X,−Y,+4 | 19 | c.2246_2249del | p.R749PfsX29 | FLT3/TKD, NPM1 |
| 27 | 75/F | M1 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 28 | 51/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 29 | 60/F | M1 | 46,XX,t(9;22)(q34;q11) | 18 | c.2113A > T | p.I705F§ | IDH1 |
| 30 | 73/F | M1 | 46,XX | 23 | c.2646G > A | p.R882H | CEBPA, TET2 |
| 31 | 22/F | M4 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 32 | 38/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 33 | 31/F | M5 | 46,XX | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 34 | 46/M | M4 | 45X,−Y | 23 | c.2645C > A | p.R882S | NRAS, FLT3/ITD, NPM1 |
| 35 | 80/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD |
| 36 | 52/M | M1 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, IDH2 |
| 37 | 44/M | M2 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 38 | 33/F | M1 | 46,XX | 8 | c.958C > T | p.R320X | FLT3/TKD, NPM1 |
| 39 | 42/M | M4 | 45,X,−Y | 15 | c.1816C > T | p.Q606X | NPM1 |
| 40 | 78/F | M2 | 46,XX | 19 | c.2255_2257del | p.F752del | FLT3/ITD, NPM1 |
| 41 | 75/F | M2 | 47,XX,del(5)(q22q35),+8 | 8 | c.958C > T | p.R320X | IDH2 |
| 42 | 49/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 43 | 78/M | M4 | 46,XY | 4 | c.315C > A | p.S105R | PTPN11 |
| 44 | 64/M | M5 | 46,XY | 23 | c.2645C > A | p.R882S | PTPN11, MLL/PTD |
| 45 | 40/M | M5 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1 |
| 46 | 75/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | NRAS, NPM1, TET2 |
| 47 | 58/F | M2 | 46,XX | 23 | c.2645C > T | p.R882C | NPM1 |
| 48 | 48/F | M1 | 47,XX,+8 | 8 | c.1001delG | p.G334AfsX11 | CEBPA, IDH2 |
| 49 | 67/M | M5 | 47,XY,+8 | 23 | c.2645C > T | p.R882C | PTPN11, KRAS, AML1/RUNX1, IDH2 |
| 50 | 51/M | M2 | 46XY | 23 | c.2646G > A | p.R882H | IDH2 |
| 51 | 85/M | M5 | 46,XY | 7 | c.767_770del | P256LfsX59 | FLT3/ITD, NPM1, WT1 |
| 52 | 85/M | M1 | 45,XY,−7 | 4 | c.327_328insG | Q110AfsX14 | |
| 53 | 67/M | M8 | 47,XY,+8 | 23 | c.2646G > A | p.R882H | NRAS, IDH2 |
| 54 | 35/M | M4 | 46,XY | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1 |
| 55 | 47/F | M2 | 46,XX | 23 | c.2646G > A | p.R882H | ASXL1, IDH2 |
| 56 | 50/F | M1 | 46,XX | 23 | c.2645C > T | p.R882C | FLT3/ITD, MLL/PTD |
| 57 | 86/M | M5 | 46,XY | 8 | c.866delG | p.G289AfsX26 | FLT3/ITD |
| 58 | 69/M | M1 | NM | 23 | c.2645C > T | p.R882C | FLT3/ITD, NPM1, CEBPA |
| 59 | 75/M | M4 | 46,XY | 23 | c.2646G > A | p.R882H | AML1/RUNX1 |
| 60 | 79/F | M4 | Cplx* | 22 | c.2510C > G | p.S837X | |
| 61 | 61/M | M1 | 46,XY | 23 | c.2646G > A | p.R882H | NPM1, WT1, TET2 |
| 62 | 37/F | M2 | 46,XX | 19 | c.2312G > A | p.R771Q‡ | NPM1, TET2 |
| 63 | 70/M | M2 | 46,XY | 23 | c.2646G > A | p.R882H | FLT3/ITD, NPM1, IDH2 |
| 64 | 46/M | M4 | 46,XY | 19 | c.2182G > C, c.2191T > C | p.G728R‡, p.F731L‡ | FLT3/ITD |
| 65 | 69/M | M4 | 47,XY,+X | 8 | c.941G > A | p.W314X | NRAS, FLT3/TKD, AML1/RUNX1, IDH2 |
| 19 | c.2207G > A | p.R736H† | |||||
| 66 | 38/F | M2 | 46,XX | 19 | c.2207G > A | p.R736H† | FLT3/ITD, NPM1, IDH1 |
| 67 | 66 | M1 | 47,XY,del(5)(q31q35), der(7)t(5;7)(q13;q11),+8 | 15 | c.1792C > T | p.R598X | IDH2 |
| 68 | 81 | M4 | 46,XY | 17 | c.2032C > T | p.Q678X | NRAS, TET2 |
| 19 | c.2210T > A | p.L737H‡ | |||||
| 69 | 50 | M4 | 46,XX | 15 | c.1903C > T | p.R635W† | PTPN11, NPM1, IDH2 |
| 70 | 84 | M0 | ND | 19 | c.2207G > A | p.R736H† | AML1/RUNX1, IDH2 |
Nucleotide numberings are according to the National Center for Biotechnology Information reference sequence NM_024426.
UPN indicates unique patient number; NM, no mitosis; and ND, not done.
Cplx: complex abnormalities, including del(2)(q31q35),der(2)del(2)(p12p22)del(2)(q31q35),−5,+6,del(7)(q11q36),+8,+11,del(12)(p11.1.p11.2),add(17)(p11).
In addition to R882 mutations, missense mutations in patients 65, 66, 69, and 70 have been reported in previous studies.4,7
Missense mutations in patients 20, 29, 62, 64, and 68 were confirmed to be significant by the analysis of remission BM samples.
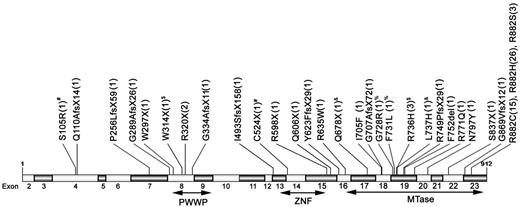
Patterns and locations of the 30 different positions of mutations. The positions and predicted translational consequences of DNMT3A mutations detected in 500 AML samples are shown. The number of patients with the mutation is indicated in the parentheses behind each mutation. #, %, &, and $ indicate that the patient has 2 mutations.
Patterns and locations of the 30 different positions of mutations. The positions and predicted translational consequences of DNMT3A mutations detected in 500 AML samples are shown. The number of patients with the mutation is indicated in the parentheses behind each mutation. #, %, &, and $ indicate that the patient has 2 mutations.
Correlation of DNMT3A mutations with clinical and laboratory features
In total, 500 de novo AML patients, including 70 (14%) DNMT3A-mutated and 430 DNMT3A-wild patients were enrolled into the study. The 6 patients with missense mutations of unknown significance were censored and were not included in the following analyses. A comparison of clinical characteristics of patients with and without distinct DNMT3A mutations is given in Table 2. DNMT3A-mutated patients were older (median, 61 vs 49 years, P < .0001) and had higher WBC, blast, and platelet counts than DNMT3A-wild patients (P = .0018, .0012, and .0001, respectively). Patients with the FAB M5 subtype of AML had the highest incidence (50%, P < .0001) of DNMT3A mutation, followed by those with the FAB M4 subtype (22.6%, P = .0026). DNMT3A mutations were positively associated with the expression of CD13 (P = .022) and CD14 (P = .0015), but inversely associated with the expression of CD34 (P = .0039) on leukemic cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). There was no difference in the expression of other antigens between the patients with and without the DNMT3A mutation.
Comparison of clinical and laboratory features between AML patients with and without the DNMT3A mutation
| Variable . | Total (n = 500) . | DNMT3A-mutated (n = 70, 14%) . | DNMT3A-wild (n = 430, 86%) . | P . |
|---|---|---|---|---|
| Sex, n | .7961 | |||
| Male | 285 | 41 | 244 | |
| Female | 215 | 29 | 186 | |
| Median age, y (range) | 51 (15-90) | 61 (16-87) | 49 (15-90) | < .0001 |
| Laboratory data, median (range) | ||||
| WBCs, /μL | 19 075 (120-627 800) | 32 490 (650-340 400) | 15 940 (120-627 800) | .0018 |
| Hemoglobin, g/dL | 8 (2.9-16.2) | 8.7 (3.2-12) | 7.9 (2.9-16.2) | .0675 |
| Platelets, × 1000/μL | 42 (2-802) | 57 (10-436) | 39 (2-802) | .0001 |
| Blasts, /μL | 7401 (0-456 725) | 19 030 (111-283 212) | 6263 (0-456 725) | .0012 |
| Lactase dehydrogenase, U/L | 889 (206-15 000) | 1064 (250-8116) | 832 (206-15 000) | .0883 |
| FAB, n (%) | < .0001 | |||
| M0 | 10 | 2 (20) | 8 (80) | .6375 |
| M1 | 112 | 14 (12.5) | 98 (87.5) | .7572 |
| M2 | 171 | 13 (7.6) | 158 (92.4) | .0027 |
| M3 | 38 | 0 (0) | 38 (100) | .0056 |
| M4 | 124 | 28 (22.6) | 96 (77.4) | .0026 |
| M5 | 24 | 12 (50) | 12 (50) | < .0001 |
| M6 | 12 | 0 (0) | 12 (100) | .3889 |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | > .9999 |
| Variable . | Total (n = 500) . | DNMT3A-mutated (n = 70, 14%) . | DNMT3A-wild (n = 430, 86%) . | P . |
|---|---|---|---|---|
| Sex, n | .7961 | |||
| Male | 285 | 41 | 244 | |
| Female | 215 | 29 | 186 | |
| Median age, y (range) | 51 (15-90) | 61 (16-87) | 49 (15-90) | < .0001 |
| Laboratory data, median (range) | ||||
| WBCs, /μL | 19 075 (120-627 800) | 32 490 (650-340 400) | 15 940 (120-627 800) | .0018 |
| Hemoglobin, g/dL | 8 (2.9-16.2) | 8.7 (3.2-12) | 7.9 (2.9-16.2) | .0675 |
| Platelets, × 1000/μL | 42 (2-802) | 57 (10-436) | 39 (2-802) | .0001 |
| Blasts, /μL | 7401 (0-456 725) | 19 030 (111-283 212) | 6263 (0-456 725) | .0012 |
| Lactase dehydrogenase, U/L | 889 (206-15 000) | 1064 (250-8116) | 832 (206-15 000) | .0883 |
| FAB, n (%) | < .0001 | |||
| M0 | 10 | 2 (20) | 8 (80) | .6375 |
| M1 | 112 | 14 (12.5) | 98 (87.5) | .7572 |
| M2 | 171 | 13 (7.6) | 158 (92.4) | .0027 |
| M3 | 38 | 0 (0) | 38 (100) | .0056 |
| M4 | 124 | 28 (22.6) | 96 (77.4) | .0026 |
| M5 | 24 | 12 (50) | 12 (50) | < .0001 |
| M6 | 12 | 0 (0) | 12 (100) | .3889 |
| Undetermined | 9 | 1 (11.1) | 8 (88.9) | > .9999 |
Association of DNMT3A mutations with cytogenetic abnormalities
Chromosome data were available for 482 patients at diagnosis, including 66 DNMT3A-mutated and 416 DNMT3A-wild patients (Table 3). DNMT3A mutations occurred more frequently in patients with intermediate-risk cytogenetics (19.5%) than in those with favorable karyotype or unfavorable cytogenetics (2.4%, P = .0069). There was a significant difference in the incidence of the DNMT3A mutation among patients with normal karyotype (22.9%), simple abnormalities with 1 or 2 changes (6.2%), and complex cytogenetics with 3 or more abnormalities (3.9%, P < .0001). None of the patients with t(8;21), t(15;17) inv(16), or 11q23 translocation had a DNMT3A mutation. There was no association of the DNMT3A mutation with other chromosomal abnormalities, including +8, +11, +13, +21, −5/del(5q), and −7/del(7q).
Association of DNMT3A mutation with chromosomal abnormalities
| . | Total . | DNMT3A-mutated . | DNMT3A-wild . | P . |
|---|---|---|---|---|
| Karyotype* | .0069 | |||
| Favorable | 99 | 0 (0) | 99 (100) | < .0001 |
| Intermediate | 318 | 62 (19.5) | 256 (80.5) | < .0001 |
| Unfavorable | 65 | 4 (6.2) | 61 (93.8) | .0783 |
| Unknown | 18 | 4 (22.2) | 14 (77.7) | |
| Normal | 223 | 51 (22.9) | 172 (77.1) | < .0001 |
| Simple | 208 | 13 (6.2) | 195 (93.8) | < .0001 |
| Complex | 51 | 2 (3.9) | 49 (96.1) | .0303 |
| t(8;21) | 42 | 0 (0) | 42 (100) | .0034 |
| t(15;17) | 38 | 0 (0) | 38 (100) | .0053 |
| inv(16) | 19 | 0 (0) | 19 (100) | .0909 |
| t(11q23) | 16 | 0 (0) | 16 (100) | .145 |
| t(7;11) | 10 | 0 (0) | 10 (100) | .371 |
| −5/5q−† | 2 | 1 (50) | 1 (50) | .2554 |
| −7/7q−† | 10 | 1 (10) | 9 (90) | > .9999 |
| +8‡ | 27 | 4 (14.8) | 23 (85.2) | .7765 |
| +11† | 3 | 1 (33.3) | 2 (66.7) | .3577 |
| +13† | 1 | 0 (0) | 1 (100) | > .9999 |
| +21† | 9 | 0 (0) | 9 (100) | .6178 |
| . | Total . | DNMT3A-mutated . | DNMT3A-wild . | P . |
|---|---|---|---|---|
| Karyotype* | .0069 | |||
| Favorable | 99 | 0 (0) | 99 (100) | < .0001 |
| Intermediate | 318 | 62 (19.5) | 256 (80.5) | < .0001 |
| Unfavorable | 65 | 4 (6.2) | 61 (93.8) | .0783 |
| Unknown | 18 | 4 (22.2) | 14 (77.7) | |
| Normal | 223 | 51 (22.9) | 172 (77.1) | < .0001 |
| Simple | 208 | 13 (6.2) | 195 (93.8) | < .0001 |
| Complex | 51 | 2 (3.9) | 49 (96.1) | .0303 |
| t(8;21) | 42 | 0 (0) | 42 (100) | .0034 |
| t(15;17) | 38 | 0 (0) | 38 (100) | .0053 |
| inv(16) | 19 | 0 (0) | 19 (100) | .0909 |
| t(11q23) | 16 | 0 (0) | 16 (100) | .145 |
| t(7;11) | 10 | 0 (0) | 10 (100) | .371 |
| −5/5q−† | 2 | 1 (50) | 1 (50) | .2554 |
| −7/7q−† | 10 | 1 (10) | 9 (90) | > .9999 |
| +8‡ | 27 | 4 (14.8) | 23 (85.2) | .7765 |
| +11† | 3 | 1 (33.3) | 2 (66.7) | .3577 |
| +13† | 1 | 0 (0) | 1 (100) | > .9999 |
| +21† | 9 | 0 (0) | 9 (100) | .6178 |
Four hundred eighty-two patients, including 66 DNMT3A-mutated and 416 DNMT3A-wild patients, had chromosome data at diagnosis.
Favorable, t(15;17), t(8;21), inv(16); unfavorable, −7, del(7q), −5, del(5q), 3q abnormality, complex abnormalities; and intermediate, normal karyotype and other abnormalities.
Only including simple chromosomal abnormalities with ≤ 2 changes, but not those with complex abnormalities with ≥ 3 aberrations.
Association of DNMT3A mutation with other molecular abnormalities
To investigate the interaction of gene mutations in the pathogenesis of adult AML, a complete mutational screening of 16 other genes was performed in all 500 patients (Table 4). Among the 70 patients with DNMT3A mutations, 68 (97.1%) showed additional molecular abnormalities at diagnosis (supplemental Table 2). Fifteen had 1 additional change, 37 had 2, 13 had 3, and 3 had 4. The most common associated molecular event was the NPM1 mutation (n = 38), followed by FLT3-ITD (n = 30), IDH2 (n = 16), and FLT3-TKD (n = 9) mutations. Patients with DNMT3A mutations had a significantly higher incidence of the NPM1 mutation and FLT3-ITD, IDH2, and PTPN11 mutations than those with DNMT3A-wild type (54.3% vs 15.3%, P < .0001; 42.9% vs 19.3%, P < .0001; 22.9% vs 9.1%, P = .0016; and 10% vs 3.5%; P = .007, respectively). Conversely, CEBPA mutation was rarely seen in patients with DNMT3A mutations (4.3% vs 14.7%, P = .0134). There was no difference in the incidence of other molecular mutations between patients with and without the DNMT3A mutation. Among the 68 patients with concurrent other genetic alterations, 51 (75%) had at least 1 concomitant class I mutation; 16 (23.5%) had class II mutations; and 38 (54.3%) had NPM1 mutations, which behave more like class II mutations.13 In total, 40 patients (58.8%) had concurrent class I and class II or NPM1 mutations at diagnosis. Twenty-one patients had concomitant FLT3-ITD and NPM1 mutations (supplemental Table 2).
Association of the DNMT3 mutation with other gene mutations
| Variable . | Patients with alteration, n (%) . | P . | ||
|---|---|---|---|---|
| Whole cohort (n = 500) . | DNMT3A-mutated patients (n = 70) . | DNMT3A-wild patients (n = 430) . | ||
| FLT3/ITD | 113 (22.6) | 30 (42.9) | 83 (19.3) | < .0001 |
| FLT3/TKD | 39 (7.8) | 9 (12.9) | 29 (6.7) | .087 |
| N-RAS | 61 (12.2) | 8 (11.4) | 53 (12.3) | > .9999 |
| K-RAS | 16 (3.2) | 1 (1.4) | 15 (3.5) | .7112 |
| PTPN11 | 18 (3.6) | 7 (10) | 11 (2.6) | .007 |
| KIT | 15 (3.0) | 0 (0) | 15 (3.5) | .2451 |
| JAK2 | 3 (0.6) | 0 (0) | 3 (0.7) | > .9999 |
| WTI | 33 (6.6) | 2 (2.9) | 31 (7.2) | .2946 |
| NPM1 | 104 (20.8) | 38 (54.3) | 66 (15.3) | < .0001 |
| CEBPA | 66 (13.2) | 3 (4.3) | 63 (14.7) | .0134 |
| AML1/RUNX1 | 62 (12.4) | 8 (11.4) | 54 (12.6) | > .9999 |
| MLL/PTD | 27 (5.4) | 6 (8.6) | 21 (4.9) | .2475 |
| ASXL1 | 51 (10.2) | 4 (5.7) | 46 (10.7) | .2812 |
| IDH1 | 27 (5.4) | 4 (5.7) | 23 (5.3) | .7812 |
| IDH2 | 55 (11) | 16 (22.9) | 39 (9.1) | .0016 |
| TET2 | 65 (13.0) | 6 (8.6) | 59 (13.7) | .3365 |
| Variable . | Patients with alteration, n (%) . | P . | ||
|---|---|---|---|---|
| Whole cohort (n = 500) . | DNMT3A-mutated patients (n = 70) . | DNMT3A-wild patients (n = 430) . | ||
| FLT3/ITD | 113 (22.6) | 30 (42.9) | 83 (19.3) | < .0001 |
| FLT3/TKD | 39 (7.8) | 9 (12.9) | 29 (6.7) | .087 |
| N-RAS | 61 (12.2) | 8 (11.4) | 53 (12.3) | > .9999 |
| K-RAS | 16 (3.2) | 1 (1.4) | 15 (3.5) | .7112 |
| PTPN11 | 18 (3.6) | 7 (10) | 11 (2.6) | .007 |
| KIT | 15 (3.0) | 0 (0) | 15 (3.5) | .2451 |
| JAK2 | 3 (0.6) | 0 (0) | 3 (0.7) | > .9999 |
| WTI | 33 (6.6) | 2 (2.9) | 31 (7.2) | .2946 |
| NPM1 | 104 (20.8) | 38 (54.3) | 66 (15.3) | < .0001 |
| CEBPA | 66 (13.2) | 3 (4.3) | 63 (14.7) | .0134 |
| AML1/RUNX1 | 62 (12.4) | 8 (11.4) | 54 (12.6) | > .9999 |
| MLL/PTD | 27 (5.4) | 6 (8.6) | 21 (4.9) | .2475 |
| ASXL1 | 51 (10.2) | 4 (5.7) | 46 (10.7) | .2812 |
| IDH1 | 27 (5.4) | 4 (5.7) | 23 (5.3) | .7812 |
| IDH2 | 55 (11) | 16 (22.9) | 39 (9.1) | .0016 |
| TET2 | 65 (13.0) | 6 (8.6) | 59 (13.7) | .3365 |
Impact of DNMT3A mutation on response to therapy and clinical outcome
Of the 363 AML patients undergoing conventional intensive induction chemotherapy, 284 (78.5%) patients achieved a CR. The probability of achieving a CR was similar between patients with and without DNMT3A mutations (74.4% vs 79%, P = .5531). However, the patients with DNMT3A mutations had a trend of higher relapse rate than those without (65.6% vs 48.8%, P = .0911). With a median follow-up of 55 months (range, 1.0-160), patients with the DNMT3A mutation had significantly poorer OS and RFS than those without the DNMT3A mutation (median, 14.5 months vs 38 months, P = .013, and median, 7.5 months vs 15 months, P = .012, respectively, Figure 2A-C). The same was true among patients with non-M3 AML (P = .04 and P = .036, respectively). In the subgroup of 130 younger patients (< 60 years) with normal karyotype AML (CN-AML), the differences between patients with and without the DNMT3A mutation in OS (median, 15.5 months vs not reached, P = .018, Figure 2B) and RFS (median, 6 months vs 21 months, P = .004, Figure 2D) were still significant. We also observed that the prognostic impact of the DNMT3A mutation could only be demonstrated in the patients with a poor prognostic genotype (NPM1-mutated (NPM1+)/FLT3-ITD+, NPM1-wild (NPM1−)/FLT3-ITD+ or FLT3/ITD−), but not in those with favorable genotype (NPM1+/FLT3-ITD−) among total AML patients (P < .001 and P = .823, respectively) or in CN-AML patients (P < .001 and P = .970, respectively). There was no significant difference in survival between patients with mutations of R882 and those with other mutations (P = .612).
OS and RFS in total patients and in younger patients with CN-AML. Kaplan-Meier survival curves for OS and RFS in 363 AML patients (A and C) and 130 younger patients (< 60 years) with CN-AML (B and D) who received standard intensive chemotherapy.
OS and RFS in total patients and in younger patients with CN-AML. Kaplan-Meier survival curves for OS and RFS in 363 AML patients (A and C) and 130 younger patients (< 60 years) with CN-AML (B and D) who received standard intensive chemotherapy.
In multivariate analysis (Table 5), the independent poor risk factors for OS were older age (> 50 years), high WBC count (> 50 000/μL), unfavorable karyotype, DNMT3A mutation, AML1/RUNX1 mutation, and WT1 mutation. Conversely, CEBPAdouble-mutation and NPM1+/FLT3−ITD− were independent favorable prognostic factors. There was a trend of better OS in patients with the IDH2 mutation (hazard ratio [HR], 0.573; 95% confidence interval [95% CI], 0.296-1.110, P = .099). The independent poor risk factors for RFS included high WBC count (> 50 000/μL), unfavorable karyotype, DNMT3A mutation, and WT1 mutation. NPM1+/FLT3-ITD− was an independent favorable factor for RFS. In 130 CN-AML patients younger than 60 years, the DNMT3A mutation was still an independent poor prognosis for OS and RFS (HR, 2.303; 95% CI, 1.088-4.876, P = .029 and HR, 3.496; 95% CI, 1.773-6.896, P < .001, respectively, supplemental Table 3).
Multivariate analysis (Cox regression) for relapse-free and overall survival
| Variable . | Relapse-free survival . | Overall survival . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||
| Lower . | Upper . | Lower . | Upper . | |||||
| Age* | 1.150 | 0.803 | 1.648 | .446 | 2.531 | 1.790 | 3.580 | < .001† |
| WBC‡ | 1.649 | 1.120 | 2.428 | .011† | 1.970 | 1.358 | 2.857 | < .001† |
| Karyotype§ | 2.577 | 1.433 | 4.633 | .002† | 3.078 | 1.849 | 5.123 | < .001† |
| NPM1/FLT3-ITD¶ | 0.268 | 0.124 | 0.581 | .001† | 0.261 | 0.121 | 0.564 | .001† |
| CEBPA# | 0.629 | 0.362 | 1.093 | .100 | 0.423 | 0.211 | 0.847 | .015† |
| IDH2** | 0.775 | 0.420 | 1.430 | .415 | 0.573 | 0.296 | 1.110 | .099 |
| WT1 | 2.823 | 1.680 | 4.743 | <.001† | 2.576 | 1.490 | 4.454 | .001† |
| AML1/RUNX1 | 1.448 | 0.718 | 2.918 | .301 | 1.963 | 1.129 | 3.414 | .017† |
| ASXL1 | 0.739 | 0.293 | 1.863 | .521 | 1.439 | 0.798 | 2.597 | .227 |
| TET2 | 1.125 | 0.625 | 2.026 | .694 | 1.033 | 0.601 | 1.777 | .906 |
| DNMT3A | 2.898 | 1.673 | 5.022 | <.001† | 2.218 | 1.333 | 3.692 | .002† |
| Variable . | Relapse-free survival . | Overall survival . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |||
| Lower . | Upper . | Lower . | Upper . | |||||
| Age* | 1.150 | 0.803 | 1.648 | .446 | 2.531 | 1.790 | 3.580 | < .001† |
| WBC‡ | 1.649 | 1.120 | 2.428 | .011† | 1.970 | 1.358 | 2.857 | < .001† |
| Karyotype§ | 2.577 | 1.433 | 4.633 | .002† | 3.078 | 1.849 | 5.123 | < .001† |
| NPM1/FLT3-ITD¶ | 0.268 | 0.124 | 0.581 | .001† | 0.261 | 0.121 | 0.564 | .001† |
| CEBPA# | 0.629 | 0.362 | 1.093 | .100 | 0.423 | 0.211 | 0.847 | .015† |
| IDH2** | 0.775 | 0.420 | 1.430 | .415 | 0.573 | 0.296 | 1.110 | .099 |
| WT1 | 2.823 | 1.680 | 4.743 | <.001† | 2.576 | 1.490 | 4.454 | .001† |
| AML1/RUNX1 | 1.448 | 0.718 | 2.918 | .301 | 1.963 | 1.129 | 3.414 | .017† |
| ASXL1 | 0.739 | 0.293 | 1.863 | .521 | 1.439 | 0.798 | 2.597 | .227 |
| TET2 | 1.125 | 0.625 | 2.026 | .694 | 1.033 | 0.601 | 1.777 | .906 |
| DNMT3A | 2.898 | 1.673 | 5.022 | <.001† | 2.218 | 1.333 | 3.692 | .002† |
HR indicates hazard ratio; and 95% CI, 95% confidence interval.
Age > 50 relative to age ≤ 50 (the reference age).
Statistically significant (P < .05).
WBCs > 50 000/μL versus < 50 000/μL.
Unfavorable cytogenetics versus others.
NPM1mut/FLT3-ITDneg versus other subtypes.
CEBPAdouble-mutation versus others.
IDH2 mutations included R140 and R172 mutations.
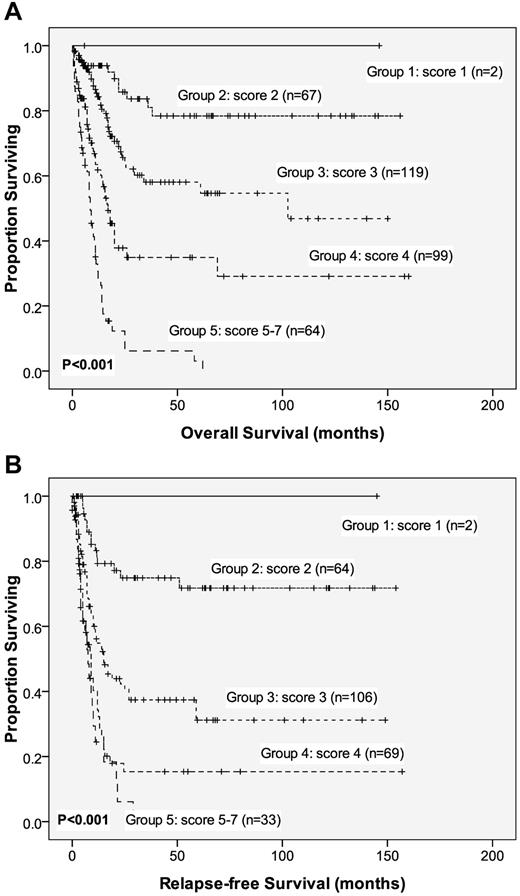
To better stratify the AML patients into different risk groups, a scoring system incorporating 9 prognostic markers—age, WBC count, cytogenetics at diagnosis, NPM1/FLT3-ITD, and mutations of CEBPA, DNMT3A, AML1/RUNX1, IDH2, and WT1 mutation—into the survival analysis was formulated based on the results of our Cox proportional hazards model. A score of −1 was assigned for each parameter associated with a favorable outcome (ie, CEBPAdouble-mutation, IDH2 mutation, and NPM1+/FLT3-ITD−), whereas a score of +1 for each factor associated with an adverse outcome (ie, DNMT3A, WT1, and AML1/RUNX1 mutations, older age, and higher WBC counts at diagnosis). The karyotypes were stratified into 3 groups (+2, unfavorable; +1, intermediate; and 0, favorable). The algebraic summation of these scores for each patient was the final score. This score system divided the AML patients into 5 groups with different clinical outcomes (P < .001 for both OS and RFS, Figure 3).
OS and RFS stratified by proposed scoring system. Kaplan-Meier survival curves for OS (A) and RFS (B) in AML patients based on our new scoring system (P < .001 for both OS and RFS). AML patients were grouped according to our scoring system based on the DNMT3A mutation and 8 other prognostic markers (ie, age, WBC count at diagnosis, and CEBPAdouble-mutation, NPM1/FLT3-ITD, IDH2, DNMT3A, WT1, and AML1/RUNX1 mutations). A score of −1 was assigned for each parameter associated with a favorable outcome (ie, CEBPAdouble-mutation, IDH2 mutation, and NPM1+/FLT3-ITD−); a score of +1 was assigned for each factor associated with an adverse outcome (ie, older age, higher WBC counts at diagnosis, and DNMT3A, WT1, and AML1/RUNX1 mutations). The karyotypes were stratified into 3 groups (+2, unfavorable; +1, intermediate; and 0, favorable). The algebraic summation of these scores for each patient was the final score. The 12 patients without chromosome data were not included in the analysis.
OS and RFS stratified by proposed scoring system. Kaplan-Meier survival curves for OS (A) and RFS (B) in AML patients based on our new scoring system (P < .001 for both OS and RFS). AML patients were grouped according to our scoring system based on the DNMT3A mutation and 8 other prognostic markers (ie, age, WBC count at diagnosis, and CEBPAdouble-mutation, NPM1/FLT3-ITD, IDH2, DNMT3A, WT1, and AML1/RUNX1 mutations). A score of −1 was assigned for each parameter associated with a favorable outcome (ie, CEBPAdouble-mutation, IDH2 mutation, and NPM1+/FLT3-ITD−); a score of +1 was assigned for each factor associated with an adverse outcome (ie, older age, higher WBC counts at diagnosis, and DNMT3A, WT1, and AML1/RUNX1 mutations). The karyotypes were stratified into 3 groups (+2, unfavorable; +1, intermediate; and 0, favorable). The algebraic summation of these scores for each patient was the final score. The 12 patients without chromosome data were not included in the analysis.
Sequential studies of DNMT3A mutations in AML patients
DNMT3A mutations were studied sequentially in 316 samples from 138 patients, including 35 patients with distinct DNMT3A mutations and 103 patients without mutations at diagnosis (Table 6). Among the 34 patients with DNMT3A mutations who had ever obtained a CR and had available samples for study, 29 lost the original mutation at remission status, but 5 (patients 5, 8, 28, 32, and 33) retained it (Table 6); all 5 patients relapsed within a median of 3.5 months and died of disease progression, suggesting the presence of leukemic cells. In the 13 patients who had available samples for serial study at relapse, 12 patients regained the original mutations, but 1 (patient 9) lost the mutation at relapse. Because direct sequencing might not be sensitive enough to detect low levels of DNMT3A mutation signal, we sequenced TA clones of the PCR product from patient 9 and 1 mutant clone of 17 was detected. Among the 103 patients who had no DNMT3A mutation at diagnosis, none acquired the DNMT3A mutation at relapse, whereas karyotypic evolution was noted at relapse in 39% of these patients (data not shown).
Sequential studies in AML patients with DNMT3A mutations
| UPN . | Date . | Status . | Karyotype . | DNMT3A mutation . | Other mutations . |
|---|---|---|---|---|---|
| 4 | 10/31/2006 | Initial | 46, XY | p.R882H | IDH2 |
| 11/29/2006 | − | ||||
| 5 | 7/27/2000 | Initial | 46,XY | p.R882C | NPM1, FLT3/TKD |
| 8/24/2000 | CR1 | p.R882C | − | ||
| 7/17/2001 | Relapse 1 | 46,XY | p.R882C | NPM1, FLT3/TKD, WT1 | |
| 10/23/2001 | CR2 | p.R882C | − | ||
| 5/7/2002 | Relapse 2 | 46,XY,del(6)(p21) | p.R882C | NPM1, FLT3/TKD, WT1 | |
| 6 | 8/31/2004 | Initial | 46,XX | p.R882H | NPM1, FLT3/ITD |
| 9/14/2006 | CR | − | − | ||
| 8 | 9/16/2005 | Initial | 46,XX,t(5;17)(q33;q21) | p.R882C | NPM1, FLT3/TKD |
| 11/4/2005 | CR | 46,XX | p.R882C | − | |
| 9 | 5/27/1997 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 6/23/1997 | CR | − | − | ||
| 7/30/1997 | Relapse | 46,XY | −* | FLT3/ITD | |
| 10 | 5/16/2000 | Initial | 46,XX | p.G707AfsX72 | NRAS, IDH1 |
| 6/7/2000 | CR | − | − | ||
| 13 | 7/26/2002 | Initial | 46,XY | p.R882H | FLT3/TKD, NPM1 |
| 9/2/2002 | CR | − | − | ||
| 14 | 12/22/2003 | Initial | 46,XY | p.Y623FfsX29 | NPM1 |
| 3/5/2004 | CR | − | − | ||
| 15 | 11/21/2006 | Initial | 46,XY | p.W297X | PTPN11, ASXL1 |
| 5/3/2007 | CR | − | − | ||
| 17 | 4/24/2007 | Initial | 46,XX | p.R882C | FLT3/TKD, AML1/RUNX1, IDH2 |
| 6/28/2007 | CR | − | − | ||
| 18 | 10/15/1999 | Initial | 46,XX | p.R882H | NRAS, NPM1 |
| 11/30/1999 | CR | − | − | ||
| 1/18/2001 | Relapse | 46,XX | p.R882H | NPM1 | |
| 20 | 12/28/2007 | Initial | 47,XX,+i(11)(q10) | p.N797Y | ASXL1, IDH2 |
| 6/20/2008 | CR | 46,XX | − | − | |
| 10/21/2008 | Relapse | 46,XX | p.N797Y | ASXL1, IDH2 | |
| 22 | 9/16/2004 | Initial | 46,XX | p.R882C | NRAS, NPM1, IDH1 |
| 10/28/2004 | CR | − | − | ||
| 28 | 8/7/2006 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 9/26/2006 | CR | p.R882H | − | ||
| 1/18/2007 | Relapse | ND | p.R882H | FLT3/ITD, NPM1 | |
| 29 | 1/27/2004 | Initial | 46,XX,t(9;22)(q34;q11) | p.I705F | IDH1 |
| 3/1/2004 | CR | 46,XX | − | − | |
| 6/9/2005 | Relapse | 46,XX,t(9;22)(q34;q11) | p.I705F | IDH1 | |
| 31 | 4/2/2001 | Initial | 46,XX | p.R882H | FLT3/ITD, NPM1 |
| 5/11/2001 | CR | − | − | ||
| 8/20/2001 | Relapse | 44-46,XX,del(20)(q11q13)[cp6]/46,XX[7] | p.R882H | FLT3/ITD, NPM1 | |
| 32 | 4/12/2000 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 7/13/2000 | CR | p.R882C | − | ||
| 10/5/2000 | Relapse | 46,XY | p.R882C | FLT3/ITD, NPM1 | |
| 33 | 10/29/2007 | Initial | 46,XX | p.R882H | FLT3/ITD, NPM1 |
| 3/18/2008 | CR | p.R882H | − | ||
| 5/8/2008 | Relapse | ND | p.R882H | FLT3/ITD, NPM1 | |
| 34 | 6/25/1998 | Initial | 45,X,-Y | p.R882S | NRAS, FLT3/ITD, NPM1 |
| 7/7/2000 | Relapse | 45,X,-Y | p.R882S | FLT3/ITD, NPM1 | |
| 8/11/2000 | CR2 | 46,XY | − | − | |
| 37 | 2/3/2006 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 4/19/2006 | CR | − | − | ||
| 5/3/2006 | Relapse | ND | p.R882C | FLT3/ITD | |
| 38 | 8/15/2002 | Initial | 46,XX | p.R320X | FLT3/TKD, NPM1 |
| 1/28/2003 | CR | − | − | ||
| 39 | 2/15/2002 | Initial | 45,X,-Y | p.Q606X | NPM1 |
| 4/8/2002 | CR | 46,XY | − | − | |
| 45 | 2/1/2005 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 3/1/2005 | CR | − | − | ||
| 11/24/2005 | Relapse | 46,XY | p.R882H | FLT3/ITD, NPM1 | |
| 47 | 6/14/2000 | Initial | 46,XX | p.R882C | NPM1 |
| 10/19/2000 | CR | − | − | ||
| 48 | 12/13/2006 | Initial | 47,XX,+8 | p.G334AfsX11 | CEBPA, IDH2 |
| 2/9/2007 | CR | − | − | ||
| 50 | 9/25/2003 | Initial | 46,XY | R882H | IDH2 |
| 6/10/2005 | CR | − | − | ||
| 51 | 5/29/2003 | Initial | 46,XY | p.P256LfsX59 | FLT3/ITD, NPM1, WT1 |
| 7/17/2003 | CR | − | − | ||
| 12/26/2003 | Relapse | ND | p.P256LfsX59 | FLT3/ITD, NPM1, WT1 | |
| 54 | 9/5/2002 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 5/28/2003 | CR | − | − | ||
| 55 | 2/21/2006 | Initial | 46,XX | p.R882H | ASXL1, IDH2 |
| 9/14/2006 | CR | − | − | ||
| 56 | 3/24/2003 | Initial | 46,XX | p.R882C | FLT3/ITD, MLL/PTD |
| 5/21/2003 | CR | − | − | ||
| 10/1/2003 | Relapse | 46,XX | p.R882C | FLT3/ITD, MLL/PTD | |
| 61 | 10/30/1995 | Initial | 46, XY | p.R882H | NPM1, WT1, TET2 |
| 1/15/1996 | CR | − | − | ||
| 10/22/1996 | Relapse | ND | p.R882H | NPM1, TET2 | |
| 62 | 9/8/1995 | Initial | 46,XX | p.R771Q | NPM1, TET2 |
| 12/19/1995 | CR | − | − | ||
| 9/23/1996 | Relapse | 46,XX | p.R771Q | NPM1, TET2 | |
| 64 | 11/2/1999 | Initial | 46,XY | p.G728R, p.F731L | FLT3/ITD |
| 3/16/2000 | CR1 | − | − | ||
| 6/12/2000 | Relapse 1 | ND | p.G728R, p.F731L | FLT3/ITD | |
| 7/14/2000 | CR2 | ND | − | − | |
| 1/11/2001 | Relapse 2 | ND | p.G728R, p.F731L | FLT3/ITD | |
| 3/13/2001 | CR3 | − | − | ||
| 66 | 3/25/2003 | Initial | 46,XX | p.R736H | FLT3/ITD, NPM1, IDH1 |
| 12/30/2003 | CR | − | − | ||
| 69 | 4/2/2001 | Initial | 46,XX | p.R635W | PTPN11, NPM1, IDH2 |
| 5/17/2001 | CR | − | − |
| UPN . | Date . | Status . | Karyotype . | DNMT3A mutation . | Other mutations . |
|---|---|---|---|---|---|
| 4 | 10/31/2006 | Initial | 46, XY | p.R882H | IDH2 |
| 11/29/2006 | − | ||||
| 5 | 7/27/2000 | Initial | 46,XY | p.R882C | NPM1, FLT3/TKD |
| 8/24/2000 | CR1 | p.R882C | − | ||
| 7/17/2001 | Relapse 1 | 46,XY | p.R882C | NPM1, FLT3/TKD, WT1 | |
| 10/23/2001 | CR2 | p.R882C | − | ||
| 5/7/2002 | Relapse 2 | 46,XY,del(6)(p21) | p.R882C | NPM1, FLT3/TKD, WT1 | |
| 6 | 8/31/2004 | Initial | 46,XX | p.R882H | NPM1, FLT3/ITD |
| 9/14/2006 | CR | − | − | ||
| 8 | 9/16/2005 | Initial | 46,XX,t(5;17)(q33;q21) | p.R882C | NPM1, FLT3/TKD |
| 11/4/2005 | CR | 46,XX | p.R882C | − | |
| 9 | 5/27/1997 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 6/23/1997 | CR | − | − | ||
| 7/30/1997 | Relapse | 46,XY | −* | FLT3/ITD | |
| 10 | 5/16/2000 | Initial | 46,XX | p.G707AfsX72 | NRAS, IDH1 |
| 6/7/2000 | CR | − | − | ||
| 13 | 7/26/2002 | Initial | 46,XY | p.R882H | FLT3/TKD, NPM1 |
| 9/2/2002 | CR | − | − | ||
| 14 | 12/22/2003 | Initial | 46,XY | p.Y623FfsX29 | NPM1 |
| 3/5/2004 | CR | − | − | ||
| 15 | 11/21/2006 | Initial | 46,XY | p.W297X | PTPN11, ASXL1 |
| 5/3/2007 | CR | − | − | ||
| 17 | 4/24/2007 | Initial | 46,XX | p.R882C | FLT3/TKD, AML1/RUNX1, IDH2 |
| 6/28/2007 | CR | − | − | ||
| 18 | 10/15/1999 | Initial | 46,XX | p.R882H | NRAS, NPM1 |
| 11/30/1999 | CR | − | − | ||
| 1/18/2001 | Relapse | 46,XX | p.R882H | NPM1 | |
| 20 | 12/28/2007 | Initial | 47,XX,+i(11)(q10) | p.N797Y | ASXL1, IDH2 |
| 6/20/2008 | CR | 46,XX | − | − | |
| 10/21/2008 | Relapse | 46,XX | p.N797Y | ASXL1, IDH2 | |
| 22 | 9/16/2004 | Initial | 46,XX | p.R882C | NRAS, NPM1, IDH1 |
| 10/28/2004 | CR | − | − | ||
| 28 | 8/7/2006 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 9/26/2006 | CR | p.R882H | − | ||
| 1/18/2007 | Relapse | ND | p.R882H | FLT3/ITD, NPM1 | |
| 29 | 1/27/2004 | Initial | 46,XX,t(9;22)(q34;q11) | p.I705F | IDH1 |
| 3/1/2004 | CR | 46,XX | − | − | |
| 6/9/2005 | Relapse | 46,XX,t(9;22)(q34;q11) | p.I705F | IDH1 | |
| 31 | 4/2/2001 | Initial | 46,XX | p.R882H | FLT3/ITD, NPM1 |
| 5/11/2001 | CR | − | − | ||
| 8/20/2001 | Relapse | 44-46,XX,del(20)(q11q13)[cp6]/46,XX[7] | p.R882H | FLT3/ITD, NPM1 | |
| 32 | 4/12/2000 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 7/13/2000 | CR | p.R882C | − | ||
| 10/5/2000 | Relapse | 46,XY | p.R882C | FLT3/ITD, NPM1 | |
| 33 | 10/29/2007 | Initial | 46,XX | p.R882H | FLT3/ITD, NPM1 |
| 3/18/2008 | CR | p.R882H | − | ||
| 5/8/2008 | Relapse | ND | p.R882H | FLT3/ITD, NPM1 | |
| 34 | 6/25/1998 | Initial | 45,X,-Y | p.R882S | NRAS, FLT3/ITD, NPM1 |
| 7/7/2000 | Relapse | 45,X,-Y | p.R882S | FLT3/ITD, NPM1 | |
| 8/11/2000 | CR2 | 46,XY | − | − | |
| 37 | 2/3/2006 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 4/19/2006 | CR | − | − | ||
| 5/3/2006 | Relapse | ND | p.R882C | FLT3/ITD | |
| 38 | 8/15/2002 | Initial | 46,XX | p.R320X | FLT3/TKD, NPM1 |
| 1/28/2003 | CR | − | − | ||
| 39 | 2/15/2002 | Initial | 45,X,-Y | p.Q606X | NPM1 |
| 4/8/2002 | CR | 46,XY | − | − | |
| 45 | 2/1/2005 | Initial | 46,XY | p.R882H | FLT3/ITD, NPM1 |
| 3/1/2005 | CR | − | − | ||
| 11/24/2005 | Relapse | 46,XY | p.R882H | FLT3/ITD, NPM1 | |
| 47 | 6/14/2000 | Initial | 46,XX | p.R882C | NPM1 |
| 10/19/2000 | CR | − | − | ||
| 48 | 12/13/2006 | Initial | 47,XX,+8 | p.G334AfsX11 | CEBPA, IDH2 |
| 2/9/2007 | CR | − | − | ||
| 50 | 9/25/2003 | Initial | 46,XY | R882H | IDH2 |
| 6/10/2005 | CR | − | − | ||
| 51 | 5/29/2003 | Initial | 46,XY | p.P256LfsX59 | FLT3/ITD, NPM1, WT1 |
| 7/17/2003 | CR | − | − | ||
| 12/26/2003 | Relapse | ND | p.P256LfsX59 | FLT3/ITD, NPM1, WT1 | |
| 54 | 9/5/2002 | Initial | 46,XY | p.R882C | FLT3/ITD, NPM1 |
| 5/28/2003 | CR | − | − | ||
| 55 | 2/21/2006 | Initial | 46,XX | p.R882H | ASXL1, IDH2 |
| 9/14/2006 | CR | − | − | ||
| 56 | 3/24/2003 | Initial | 46,XX | p.R882C | FLT3/ITD, MLL/PTD |
| 5/21/2003 | CR | − | − | ||
| 10/1/2003 | Relapse | 46,XX | p.R882C | FLT3/ITD, MLL/PTD | |
| 61 | 10/30/1995 | Initial | 46, XY | p.R882H | NPM1, WT1, TET2 |
| 1/15/1996 | CR | − | − | ||
| 10/22/1996 | Relapse | ND | p.R882H | NPM1, TET2 | |
| 62 | 9/8/1995 | Initial | 46,XX | p.R771Q | NPM1, TET2 |
| 12/19/1995 | CR | − | − | ||
| 9/23/1996 | Relapse | 46,XX | p.R771Q | NPM1, TET2 | |
| 64 | 11/2/1999 | Initial | 46,XY | p.G728R, p.F731L | FLT3/ITD |
| 3/16/2000 | CR1 | − | − | ||
| 6/12/2000 | Relapse 1 | ND | p.G728R, p.F731L | FLT3/ITD | |
| 7/14/2000 | CR2 | ND | − | − | |
| 1/11/2001 | Relapse 2 | ND | p.G728R, p.F731L | FLT3/ITD | |
| 3/13/2001 | CR3 | − | − | ||
| 66 | 3/25/2003 | Initial | 46,XX | p.R736H | FLT3/ITD, NPM1, IDH1 |
| 12/30/2003 | CR | − | − | ||
| 69 | 4/2/2001 | Initial | 46,XX | p.R635W | PTPN11, NPM1, IDH2 |
| 5/17/2001 | CR | − | − |
The results of serial studies in 103 patients without DNMT3A mutation at diagnosis are not shown in this table. None of these 103 patients acquired DNMT3A mutation at relapse.
UPN indicates unique patient number; CR, complete remission; –, negative; ND not done; and NM, no mitosis.
Using the more sensitive TA cloning technique, 1 of 17 clones showed DNMT3A mutation.
Discussion
In the present study, we found that the DNMT3A mutation was associated with distinct clinical and biologic features and was a poor prognostic factor in AML patients independent of age, WBC counts, karyotype, and other genetic markers.
DNMT3A mutations at 30 different positions, most commonly in the MTase domain, were demonstrated (Figure 1). All of the nonsense, frame-shift, and in-frame mutations generated truncated peptide with complete or partial deletion of the MTase domain and were thought to abolish the catalytic activity of this enzyme. The missense R882 mutations, the most common DNMT3A mutations, resulted in impaired enzyme activity,4,9 but the influence of other missense mutations on the enzyme remains unclear. These missense mutations all involved amino acid residues well conserved through evolution. We censored 6 patients with missense variants of unknown significance and did not include them in the analyses because there were no available remission BM samples or normal tissues to verify that their DNMT3A variants were true somatic mutations. In contrast to the report of Thol et al,3 who only found mutations between exons 15 and 23, 10 mutations in our patients were located outside of this region (Figure 1). Nine of these mutations were frame-shift or nonsense mutations and were expected to impair enzyme activity. Similar to our finding, Ley et al also detected mutations outside of exons 15 to 23.4 In the study by Thol et al, all 23 exons of DNMT3 were initially sequenced in 40 patients.3 Because only mutations between exons 15 and 23 were found in these patients, they subsequently sequenced exons 15 to 23, but not other exons, in other patients. Because all but one mutation outside of exons 15 to 23 in our study were detected in only one patient each (an incidence of 1 in 500 for each mutation), the absence of mutation in this area in 40 patients might not mean that it would not happen in other patients.
In this study, DNMT3A mutations were found in 14%, 15.2%, 19.5%, and 22.9%, respectively, in whole cohort, non-M3 AML, intermediate-risk cytogenetics, and CN-AML groups, lower than the reports of Ley et al (22.1% for total patients, 33.7% for those with intermediate-risk cytogenetics, and 36.7% for CN-AML patients)4 and Thol et al (17.8% in non-M3 and 27.2% in CN-AML patients).3 In a study of Chinese AML patients by Yan et al, DNMT3A mutations were detected in 20.5% and 13.6%, respectively, of patients with the FAB M5 and M4 subtypes of AML, but none of the patients with FAB M1 or M2 subtypes had the mutation, leading to an overall incidence of 9% for the DNMT3A mutation in the entire group of AML patients.9 Yamashita et al also reported a low incidence (4.1%) of DNMT3A mutations in Japanese AML patients.8 The reason for the variability in the incidence of DNMT3A mutations in different studies is unknown, but may be because of the differences in ethnic background, patient populations recruited, and methods used. Whether DNMT3A mutations occur less frequently in Asian than in Western AML patients needs to be determined by further studies.
In our comprehensive analysis of the 17 gene mutations in 500 patients, we found that the DNMT3A mutation was the third commonest recurrent genetic alteration, followed by FLT3-ITD and NPM1 mutations, in AML patients. In addition to its close association with NPM1 mutations and FLT3-ITD, which has been shown previously,3,4 we demonstrated herein that DNMT3A mutations were also positively associated with PTPN11 and IDH2 mutations and negatively associated with the CEBPA mutation. More intriguingly, we found that the DNMT3A mutation rarely occurred alone; all but 2 patients with DNMT3A mutations showed concurrent mutations of other genes, more frequently class I (51 of 68, 75%), but also class II mutations (16 of 68, 23.5%) and NPM1 mutations (38 of 68, 54.3%, supplemental Table 2), which behave more like class II mutations.13 In short, the development of AML may require concerted interaction among different genetic alterations.
The stability of DNMT3A mutations in the evolution of AML remains unclear. In a serial study of 5 patients with DNMT3A mutations at diagnosis, Thol et al found that the mutations disappeared at CR and reappeared at relapse in one patient tested.3 To the best of our knowledge, the present study recruited the largest number of AML patients for sequential analysis of DNMT3A mutations during the clinical course. In contrast to the instability of FLT3-ITD during disease evolution, we found that the DNMT3A mutation seemed to be stable, analogous to NPM1 and IDH1/2 mutations.13,21,22 At relapse, all DNMT3A-mutated patients who had available samples for serial study regained the same mutations, including the one in whom the mutation could be detected by a sensitive gene-cloning technique, but not by direct sequencing. Conversely, all 103 patients without DNMT3A mutation at diagnosis remained DNMT3A-wild at relapse. These results suggested that although DNMT3A mutations are important for the development of AML, they may play little role in disease progression. Given the stability of the DNMT3A mutation during AML evolution, it may be a potential biomarker for monitoring minimal residual disease.
We found that AML patients with DNMT3A mutations had distinct clinical and laboratory characteristics and a poor prognosis. Recently, many gene mutations have been detected in AML and some found to be independent prognostic factors. In the present study, to better stratify AML patients into different risk groups, a survival scoring system incorporating the DNMT3A mutation and 8 other prognostic factors, including age, WBC count, cytogenetics, and NPM1/FLT3-ITD, CEBPA, AML1/RUNX1, WT1, and IDH2 mutations, into the survival analysis was formulated. This scoring system was found to be more powerful than any single marker at separating patients into different prognostic groups. However, further study with an independent cohort will be needed to validate the proposed scoring system.
In summary, this study demonstrated that DNMT3A mutations could be detected in a substantial number of patients with de novo AML and were closely associated with older age and FAB M4/M5 subtypes. DNMT3A mutations occurred more frequently in patients with intermediate-risk cytogenetics and normal karyotype. They were mutually exclusive with CEBPA mutation, but were closely associated with FLT3/ITD, NPM1, PTPN11, and IDH2 mutations. Furthermore, the DNMT3A mutation was an independent poor risk factor for OS and RFS among total cohort and CN-AML patients. Sequential study during the clinical course showed that the DNMT3A mutation was stable during AML evolution. We conclude that the incorporation of the DNMT3A mutation with 8 other prognostic factors into survival analyses can better stratify AML patients into different risk groups.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially sponsored by grants from the National Science Council (NSC 97-2314-B002-015-MY3, 99-2314-B-002-143, 100-2325-B-002-032, and 100-2628-B-002-003-MY3) and the Department of Health (DOH100-TD-C-111-001), Taiwan, Republic of China, and the Department of Medical Research (NTUH.99P14 and 100P07), National Taiwan University Hospital, Taipei, Taiwan.
Authorship
Contribution: H.-A.H. collected the literature, managed and interpreted the data, performed the statistical analysis, and wrote the manuscript; Y.-Y.K. collected the literature, managed and interpreted the data, and wrote the manuscript; C.-Y.L. performed and interpreted the statistical analysis; L.-I.L. performed and interpreted the mutation analysis; C.-Y.C., W.-C.C., M.Y., S.-Y.H., J.-L.T., B.-S.K., S.-C.H., S.-J.W., W.T., and Y.-C.C. contributed patient samples and clinical data; M.C.L., M.-H.T., C.-F.H., Y.-C.C., C.-Y. L., F.-Y.L., and M.-C.L. performed the gene mutation and chromosomal studies; and H.-F.T. planned, designed, and coordinated the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, MD, PhD, Department of Internal Medicine, National Taiwan University Hospital, 7 Chung-Shan Street, Taipei, Taiwan; e-mail: hftien@ntu.edu.tw.
References
Author notes
H.-A.H. and Y.-Y.K. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal