We aimed to determine the prognostic impact of monosomal karyotype (MK) in acute myeloid leukemia (AML) in the context of the current World Health Organization (WHO) classification and to evaluate the outcome of MK+ patients after allogeneic HSCT. Of 1058 patients with abnormal cytogenetics, 319 (30%) were MK MK+. MK+ patients were significantly older (P = .0001), had lower white blood counts (P = .0006), and lower percentages of BM blasts (P = .0004); MK was associated with the presence of −5/5q−, −7, 7q−, abnl(12p), abnl(17p), −18/18q−, −20/20q−, inv(3)/t(3;3), complex karyotype (CK), and myelodysplasia (MDS)–related cytogenetic abnormalities (P < .0001, each); and NPM1 mutations (P < .0001), FLT3 internal tandem duplications (P < .0001), and tyrosine kinase domain mutations (P = .02) were less frequent in MK+. Response to induction therapy and overall survival in MK+ patients were dismal with a complete remission rate of 32.5% and a 4-year survival of 9%. MK retained its prognostic impact in AML with CK, AML with MDS-related cytogenetic abnormalities, and in a revised definition (MK-R) excluding cases with recurrent genetic abnormalities according to WHO classification and those with derivative chromosomes not leading to true monosomies. In younger patients, allogeneic HSCT from matched related and unrelated donors resulted in a limited improvement of overall survival.
Introduction
Using metaphase cytogenetics, an abnormal karyotype can be detected in ∼ 55% of adult acute myeloid leukemia (AML) patients.1 Based on the current World Health Organization (WHO) classification, more than two-thirds2 of AML can be categorized on their underlying cytogenetic or molecular genetic abnormalities.3 These genetic abnormalities are the most important factors in determining response to chemotherapy as well as outcome in AML.2,4,5
According to cytogenetic abnormalities, AML are currently categorized into 3 risk groups, favorable, intermediate, and adverse; the latter subgroup includes AML with complex karyotype (CK) defined by 3 or more abnormalities (in the absence of cytogenetic abnormalities listed under the WHO category “AML with recurrent genetic abnormalities”).3,6 Recently, a new cytogenetic category was introduced, that is, the monosomal karyotype (MK) defined by the presence of one single autosomal monosomy (AM; excluding isolated loss of X or Y) in association with at least one additional AM or one structural chromosomal abnormality (in the absence of core-binding factor [CBF] AML and acute promyelocytic leukemia [APL]).7 This MK category was reported to be associated with a dismal prognosis and to add prognostic information even in patients exhibiting a CK.7
The incidence and prognostic impact of MK have not yet been determined in the context of molecular markers. The objectives of our study were to evaluate the characteristics and clinical impact of MK in a large cohort of adult AML patients treated within prospective multicenter treatment trials. In particular, we were interested in studying MK in the context of the current WHO classification,3 and to evaluate the impact of allogeneic HSCT in this subset of patients.
Methods
Patients
In total, 3172 adult AML patients (median age, 54.5 years; range: 16-85 years) were enrolled on 6 prospective multicenter treatment trials of the German-Austrian AML Study Group (AMLSG) trials between 1993 and 2008. All patients received age- and response-adapted intensive induction and consolidation therapy as previously described (AML HD938 ; APL959 ; AML HD98A10 ; AML HD98B11 ; AMLSG 06-04, NCT00151255; AMLSG 07-04; NCT00151242). The studies were approved by the institutional review boards of the participating centers. All patients gave informed consent to pretreatment cytogenetic and molecular genetic analyses as well as to treatment within the prospective trials according to the Declaration of Helsinki. The diagnosis of AML was based on French-American-British Cooperative Group criteria12 for the trials AML HD93, APL95, AML HD98A, and AML HD98B, and, after 2004, on WHO 2001 criteria13 for the trials AMLSG 07-04 and AMLSG 06-04.
Cytogenetic and molecular genetic analysis
All leukemia samples were studied centrally in the reference laboratories of the AMLSG at the University of Ulm and at Hannover Medical School. Chromosome banding was performed using standard techniques, and karyotypes were described according to the International System for Human Cytogenetic Nomenclature.14 CK was defined as 3 or more chromosome abnormalities; myelodysplasia (MDS)–related cytogenetic abnormalities included those listed in the current WHO classification.3 For both categories, the absence of one of the chromosomal abnormalities including t(v;11)(v;q23) or molecular abnormalities listed under the WHO category “AML with recurrent genetic abnormalities” was a prerequisite.3,6 Three patients [n = 2, t(2;11)(p21;q23); n = 1, t(11;16)(q23;p13.3)] were listed in the group t(v;11)(v;q23) but also in the category MDS-related cytogenetic abnormalities. The category “MK” was defined as previously described.7 In addition, we evaluated a category “MK-revised” (MK-R) in which other recurrent genetic abnormalities were excluded, that were, AML with t(9;11)(p22;q23), MLLT3-MLL; t(v;11)(v;q23), other translocations involving MLL; AML with inv(3)(q21q26.2) or t(3;3)(q21;q26.2), RPN1-EVI1; AML with t(6;9)(p23;q34), DEK-NUP21. MK-R also excluded cases with derivative chromosomes not leading to true monosomies in the International Standing Committee on Human Cytogenetic Nomenclature (ISCN) karyotype designation (n = 13; supplemental Table A1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Cytogenetic abnormalities were categorized into favorable-, intermediate-, and adverse-risk group according to the European LeukemiaNet (ELN) criteria.6
Leukemia samples were analyzed for mutations in the FLT3 (FLT3 internal tandem duplication [ITD], n = 2726; FLT3 tyrosine kinase domain [TKD] mutations at codons D835 and I836, n = 2562), NPM1 (n = 2620), MLL (partial tandem duplication [PTD], n = 1966), as well as CEBPA (n = 1805) genes as previously described.15
Statistical analyses
The definition of complete remission (CR), therapeutic failures, overall survival (OS), and relapse-free survival (RFS) followed recommended criteria.16 Pairwise comparisons between patient characteristics (covariates) were performed by Mann-Whitney U or Kruskal-Wallis test for continuous variables and by Fisher exact test for categorical variables. Logistic regression models were fitted to identify factors predictive for the remission status after induction therapy. The Kaplan-Meier method was used to estimate the distribution of RFS and OS.17 Confidence interval (CI) estimation for the survival curves was based on the cumulative hazard function using the Greenwood formula for the SE estimation. Log-rank tests were used to compare survival curves between groups. The effect of allogeneic HSCT on OS as a time-dependent intervening event was estimated by using the Mantel-Byar method.18 A Cox model was used to identify prognostic variables.19 RFS analysis included only patients attaining CR after induction therapy. Prognostic models for survival were stratified by age group (age < 61 years vs age ≥ 61 years) because of different dose intensities in the treatment protocols for younger and older patients. We imputed missing data for covariates by using 10 multiple imputations in chained equations after 10 burn-in iterations incorporating predictive mean matching.20 All statistical analyses were performed with the statistical software environment R Version 2.13.0, using the R packages rms Version 3.3-1.21
Results
Frequency of cytogenetic and molecular genetic abnormalities
Cytogenetic analysis was successful in 2851 (90%) of 3172 AML. A total of 1493 (52%) of the 2851 cases had an abnormal karyotype; after exclusion of isolated losses of sex chromosomes (n = 23), CBF-AML (n = 306), and APL (n = 106), 1058 cytogenetically abnormal cases were considered for further analysis. A total of 319 (30%) of the 1058 cytogenetically abnormal AML exhibited a MK, 335 (32%) a CK, and 503 (48%) had MDS-related cytogenetic abnormalities. Of the 319 MK+ AML, 242 (76%) had CK, and 265 (83%) had MDS-related changes. All but 5 MK+ cases belonged to the cytogenetic adverse-risk category. A total of 259 (81%) of the 319 MK+ AML fulfilled the criteria of “MK-R,” that is, the MK+ category excluding (1) AML with recurrent genetic abnormalities (n = 47), and (2) cases with derivative chromosomes not leading to true monosomies (n = 13) in the karyotype designation (supplemental Table A1).
Among MK+ cases, the most frequent chromosome abnormalities were (in order of decreasing frequency): −5 or 5q− (55%), −7 (45%), abnl(17p) (41%), abnl(12p) (24%), −20 or 20q− (19%), −18 or 18q− (19%), 7q− (17%), +8 or +8q (14%), inv(3) or t(3;3) (11%), +21 or +21q (7%), +11 or +11q (6%), +22 or +22q (4%), and +13 or +13q (3%; Table 1). MK+ cases harbored the following recurrent genetic abnormalities defined by the current WHO classification3 : inv(3) or t(3;3), n = 34 (11%); t(6;9), n = 3 (1%); t(v;11)(v;q23) n = 7 (2%) [no case of t(9;11)]; mutated NPM1, n = 3 (1%).
Genetic characteristics for the subset of cytogenetically abnormal patients (n = 1058)
| Characteristics . | MK−, n (%) . | MK+, n (%) . | P . |
|---|---|---|---|
| Genetic group | |||
| Risk category* | |||
| Intermediate | 453 (61) | 5 (2) | < .0001 |
| Adverse | 286 (39) | 314 (98) | |
| Cytogenetic abnormalities | |||
| t(9;11) | 55 (7) | 0 (0) | < .0001 |
| t(v;11)(v;q23) | 54 (7) | 7 (2) | .0008 |
| t(6;9) | 16 (2) | 3 (1) | .21 |
| inv(3) or t(3;3) | 11 (2) | 34 (11) | < .0001 |
| −5 or 5q− | 61 (8) | 174 (55) | < .0001 |
| −7 | 42 (6) | 142 (45) | < .0001 |
| 7q− | 54 (7) | 54 (17) | < .0001 |
| +8 or +8q | 205 (28) | 45 (14) | < .0001 |
| +11 or +11q | 37 (5) | 20 (6) | .46 |
| abnl(12p) | 45 (6) | 77 (24) | < .0001 |
| +13 or +13q | 33 (5) | 10 (3) | .40 |
| abnl(17p) | 18 (2) | 130 (41) | < .0001 |
| −18 or 18q− | 3 (0.4) | 61 (19) | < .0001 |
| −20 or 20q− | 32 (4) | 61 (19) | < .0001 |
| +21 or +21q | 49 (7) | 23 (7) | .79 |
| +22 or +22q | 18 (2) | 13 (4) | .17 |
| MDS-related cytogenetic changes† | 238 (32) | 265 (83) | < .0001 |
| Complex karyotype* | 93 (13) | 242 (76) | < .0001 |
| Molecular genetic abnormalities | |||
| NPM1 mutation | 59 (10) | 3 (1) | < .0001 |
| No. missing | 161 | 66 | |
| FLT3-ITD | 102 (16) | 10 (4) | < .0001 |
| No. missing | 118 | 47 | |
| FLT3-TKD | 36 (7) | 6 (2) | .02 |
| No. missing | 181 | 68 |
| Characteristics . | MK−, n (%) . | MK+, n (%) . | P . |
|---|---|---|---|
| Genetic group | |||
| Risk category* | |||
| Intermediate | 453 (61) | 5 (2) | < .0001 |
| Adverse | 286 (39) | 314 (98) | |
| Cytogenetic abnormalities | |||
| t(9;11) | 55 (7) | 0 (0) | < .0001 |
| t(v;11)(v;q23) | 54 (7) | 7 (2) | .0008 |
| t(6;9) | 16 (2) | 3 (1) | .21 |
| inv(3) or t(3;3) | 11 (2) | 34 (11) | < .0001 |
| −5 or 5q− | 61 (8) | 174 (55) | < .0001 |
| −7 | 42 (6) | 142 (45) | < .0001 |
| 7q− | 54 (7) | 54 (17) | < .0001 |
| +8 or +8q | 205 (28) | 45 (14) | < .0001 |
| +11 or +11q | 37 (5) | 20 (6) | .46 |
| abnl(12p) | 45 (6) | 77 (24) | < .0001 |
| +13 or +13q | 33 (5) | 10 (3) | .40 |
| abnl(17p) | 18 (2) | 130 (41) | < .0001 |
| −18 or 18q− | 3 (0.4) | 61 (19) | < .0001 |
| −20 or 20q− | 32 (4) | 61 (19) | < .0001 |
| +21 or +21q | 49 (7) | 23 (7) | .79 |
| +22 or +22q | 18 (2) | 13 (4) | .17 |
| MDS-related cytogenetic changes† | 238 (32) | 265 (83) | < .0001 |
| Complex karyotype* | 93 (13) | 242 (76) | < .0001 |
| Molecular genetic abnormalities | |||
| NPM1 mutation | 59 (10) | 3 (1) | < .0001 |
| No. missing | 161 | 66 | |
| FLT3-ITD | 102 (16) | 10 (4) | < .0001 |
| No. missing | 118 | 47 | |
| FLT3-TKD | 36 (7) | 6 (2) | .02 |
| No. missing | 181 | 68 |
With respect to the distribution of monosomies in MK, 137 (43%) cases had one AM, 67 (21%) had 2 AM, and 115 (36%) had 3 or more AM.
Presenting clinical, cytogenetic, and molecular genetic features of MK+ patients
Patients with MK+ were significantly older compared with MK− patients (median age 58.3 vs 55.5 years; P = .0001). MK+ was associated with lower hemoglobin levels (P = .0007), lower median white blood counts (P = .0006), and lower percentages of blasts in the BM (P = .0004) as well as in peripheral blood (P = .03; Table 2).
Clinical characteristics for the subset of cytogenetically abnormal patients (n = 1058)
| Characteristics . | MK−, n (%) . | MK+, n (%) . | P . |
|---|---|---|---|
| No. | 739 | 319 | |
| Sex | .99 | ||
| Male | 406 (55) | 176 (55) | |
| Female | 333 (45) | 143 (45) | |
| Type of AML | .07 | ||
| De novo | 602 (82) | 255 (80) | |
| Secondary | 61 (8) | 19 (6) | |
| Therapy related | 68 (9) | 43 (13) | |
| Age, y | .0001 | ||
| Median (range) | 55.5 (16.7-84.5) | 58.3 (19-81.2) | |
| ≤ 60 | 488 (66) | 189 (59) | .03 |
| > 60 | 251 (34) | 130 (41) | |
| WBC, ×109/L | .0006 | ||
| Median (range) | 9.4 (0.5-427) | 5.8 (0.3-533) | |
| No. missing | 18 | 5 | |
| Hemoglobin, g/dL | .0007 | ||
| Median (range) | 9.2 (4.3-20.6) | 8.8 (3.4-14) | |
| No. missing | 21 | 5 | |
| Platelet count, ×109/L | .08 | ||
| Median | 56 (2-933) | 51 (4-916) | |
| No. missing | 20 | 5 | |
| Percentage of PB blasts | .03 | ||
| Median (range) | 31 (0-100) | 25 (0-99) | |
| No. missing | 72 | 25 | |
| Percentage of BM blasts | .0004 | ||
| Median (range) | 78.5 (4-100) | 63 (8-100) | |
| No. missing | 77 | 33 | |
| LDH value, U/L | .31 | ||
| Median (range) | 386.5 (57-6907) | 377.5 (40-5406) | |
| No. missing | 39 | 9 |
| Characteristics . | MK−, n (%) . | MK+, n (%) . | P . |
|---|---|---|---|
| No. | 739 | 319 | |
| Sex | .99 | ||
| Male | 406 (55) | 176 (55) | |
| Female | 333 (45) | 143 (45) | |
| Type of AML | .07 | ||
| De novo | 602 (82) | 255 (80) | |
| Secondary | 61 (8) | 19 (6) | |
| Therapy related | 68 (9) | 43 (13) | |
| Age, y | .0001 | ||
| Median (range) | 55.5 (16.7-84.5) | 58.3 (19-81.2) | |
| ≤ 60 | 488 (66) | 189 (59) | .03 |
| > 60 | 251 (34) | 130 (41) | |
| WBC, ×109/L | .0006 | ||
| Median (range) | 9.4 (0.5-427) | 5.8 (0.3-533) | |
| No. missing | 18 | 5 | |
| Hemoglobin, g/dL | .0007 | ||
| Median (range) | 9.2 (4.3-20.6) | 8.8 (3.4-14) | |
| No. missing | 21 | 5 | |
| Platelet count, ×109/L | .08 | ||
| Median | 56 (2-933) | 51 (4-916) | |
| No. missing | 20 | 5 | |
| Percentage of PB blasts | .03 | ||
| Median (range) | 31 (0-100) | 25 (0-99) | |
| No. missing | 72 | 25 | |
| Percentage of BM blasts | .0004 | ||
| Median (range) | 78.5 (4-100) | 63 (8-100) | |
| No. missing | 77 | 33 | |
| LDH value, U/L | .31 | ||
| Median (range) | 386.5 (57-6907) | 377.5 (40-5406) | |
| No. missing | 39 | 9 |
AML indicates acute myeloid leukemia; LDH, serum lactate dehydrogenase; MK, monosomal karyotype; PB, peripheral blood; and WBC, white blood count. Percentages may not add to 100 because of rounding.
MK+ AML were significantly associated with inv(3) or t(3;3), −5 or 5q−, −7, 7q−, abnl(12p), abnl(17p), −18 or 18q−, −20 or 20q−, complex karyotypes, and MDS-related cytogenetic changes (P < .0001, each), and they did not or less frequently exhibited t(9;11) (P < .0001), t(v;11)(v;q23) (P = .0008), and +8 or +8q (P < .0001; Table 1). Regarding molecular abnormalities, NPM1 mutations (P < .0001), as well as FLT3-ITD (P < .0001) and FLT3-TKD mutations (P = .02) were significantly less frequent in MK+ AML (Table 1). The categorization according to MK and CK resulted in 4 groups: (1) neither MK nor CR; (2) MK sole; (3) CK sole; and (4) presence of both MK as well as CK (Table 3). The distribution of cytogenetic abnormalities according to these 4 groups revealed 3 patterns, (1) presence of the specific abnormality in all subgroups and highest incidence in the group defined as CK and MK (−5 or 5q−, 7q−, abnl(12p), abnl(17p), −18 or 18q−, −20 or 20q−); (2) highest incidence in CK sole and no or only few cases in MK sole (+8 or +8q, +11 or +11q, +13 or +13q, +21 or +21q, +22 or +22q); (3) highest incidence in MK sole and only few cases in CK sole (−7). A large proportion of MK sole cases exhibited an inv(3) or t(3;3), which was frequently associated with monosomy 7. Categorization according to MK and MDS-related cytogenetic abnormalities resulted again in 4 groups showing similarly high incidences of specific abnormalities (−5 or 5q−, 7q−, abnl(12p), abnl(17p), −18 or 18q−, −20 or 20q−) in the subgroups defined by MDS-related cytogenetic abnormalities and MK (Table 4).
Genetic characteristics according to complex and monosomal karyotype (n = 1058)
| Characteristics/cytogenetic abnormalities . | Other, n (%), n = 646 . | MK+, n (%), n = 77 . | CK+, n (%), n = 93 . | MK+CK+, n (%), n = 242 . | P . |
|---|---|---|---|---|---|
| t(9;11) | 55 (9) | – | – | – | † |
| t(v;11)(v;q23)* | 52 (8) | 7 (9) | – | – | † |
| t(6;9)* | 16 (3) | 3 (4) | – | – | † |
| inv(3) or t(3;3) | 11 (2) | 34 (44) | – | – | † |
| −5 or 5q− | 34 (5) | 8 (10) | 27 (29) | 166 (69) | < .0001 |
| −7 | 41 (6) | 57 (74) | 1 (1) | 85 (35) | < .0001 |
| 7q− | 41 (6) | 7 (9) | 13 (14) | 47 (19) | < .0001 |
| +8 or +8q | 164 (25) | 3 (4) | 41 (44) | 42 (17) | < .0001 |
| +11 or +11q | 23 (4) | 0 | 14 (15) | 20 (8) | < .0001 |
| abnl(12p) | 30 (5) | 5 (7) | 15 (16) | 72 (30) | < .0001 |
| +13 or +13q | 23 (4) | 0 | 10 (11) | 10 (4) | .005 |
| abnl(17p) | 9 (1) | 7 (9) | 9 (10) | 123 (51) | < .0001 |
| −18 or 18q− | 1 (0.2) | 6 (8) | 2 (2) | 55 (23) | < .0001 |
| −20 or 20q− | 25 (4) | 7 (9) | 7 (8) | 54 (22) | < .0001 |
| +21 or +21q | 31 (5) | 1 (1) | 18 (19) | 22 (9) | < .0001 |
| +22 or +22q | 5 (1) | 0 | 13 (14) | 13 (5) | < .0001 |
| Characteristics/cytogenetic abnormalities . | Other, n (%), n = 646 . | MK+, n (%), n = 77 . | CK+, n (%), n = 93 . | MK+CK+, n (%), n = 242 . | P . |
|---|---|---|---|---|---|
| t(9;11) | 55 (9) | – | – | – | † |
| t(v;11)(v;q23)* | 52 (8) | 7 (9) | – | – | † |
| t(6;9)* | 16 (3) | 3 (4) | – | – | † |
| inv(3) or t(3;3) | 11 (2) | 34 (44) | – | – | † |
| −5 or 5q− | 34 (5) | 8 (10) | 27 (29) | 166 (69) | < .0001 |
| −7 | 41 (6) | 57 (74) | 1 (1) | 85 (35) | < .0001 |
| 7q− | 41 (6) | 7 (9) | 13 (14) | 47 (19) | < .0001 |
| +8 or +8q | 164 (25) | 3 (4) | 41 (44) | 42 (17) | < .0001 |
| +11 or +11q | 23 (4) | 0 | 14 (15) | 20 (8) | < .0001 |
| abnl(12p) | 30 (5) | 5 (7) | 15 (16) | 72 (30) | < .0001 |
| +13 or +13q | 23 (4) | 0 | 10 (11) | 10 (4) | .005 |
| abnl(17p) | 9 (1) | 7 (9) | 9 (10) | 123 (51) | < .0001 |
| −18 or 18q− | 1 (0.2) | 6 (8) | 2 (2) | 55 (23) | < .0001 |
| −20 or 20q− | 25 (4) | 7 (9) | 7 (8) | 54 (22) | < .0001 |
| +21 or +21q | 31 (5) | 1 (1) | 18 (19) | 22 (9) | < .0001 |
| +22 or +22q | 5 (1) | 0 | 13 (14) | 13 (5) | < .0001 |
CK indicates complex karyotype; MK, monosomal karyotype; and –, not applicable. Percentages may not add to 100 because of rounding.
One case exhibited a t(6;9) and t(v;11)(v;q23) concurrently.
Genetic characteristics according to the WHO classification category AML with MDS-related cytogenetic abnormalities and MK (n = 1058)
| Characteristics/cytogenetic abnormalities . | Other, n (%), n = 501 . | MK+, n (%), n = 54 . | MDS+, n (%), n = 238 . | MK+MDS+, n (%), n = 265 . | P . |
|---|---|---|---|---|---|
| t(9;11) | 55 (11) | – | – | – | † |
| t(v;11)(v;q23)* | 52 (10) | 6 (11) | 2 (1) | 1 | < .0001 |
| t(6;9)* | 16 (3) | 3 (6) | – | – | † |
| inv(3) or t(3;3) | 11 (2) | 34 (63) | – | – | † |
| −5 or 5q− | 8 (2) | 5 (9) | 53 (22) | 169 (64) | < .0001 |
| −7 | 36 (67) | 42 (18) | 106 (40) | < .0001 | |
| 7q− | 14 (3) | 5 (9) | 40 (17) | 49 (18) | < .0001 |
| +8 or +8q | 157 (31) | 3 (6) | 48 (20) | 42 (16) | < .0001 |
| +11 or +11q | 23 (5) | 0 | 14 (6) | 20 (8) | < .0001 |
| abnl(12p) | 18 (4) | 4 (7) | 27 (11) | 73 (28) | < .0001 |
| +13 or +13q | 21 (4) | 0 | 12 (5) | 10 (4) | .44 |
| abnl(17p) | 6 (1) | 6 (11) | 12 (5) | 124 (47) | < .0001 |
| −18 or 18q− | 1 (0.2) | 5 (9) | 2 (1) | 56 (21) | < .0001 |
| −20 or 20q− | 23 (5) | 4 (7) | 9 (4) | 57 (22) | < .0001 |
| +21 or +21q | 28 (6) | 0 | 21 (9) | 23 (9) | < .0001 |
| +22 or +22q | 4 (1) | 0 | 14 (6) | 13 (5) | < .0001 |
| Characteristics/cytogenetic abnormalities . | Other, n (%), n = 501 . | MK+, n (%), n = 54 . | MDS+, n (%), n = 238 . | MK+MDS+, n (%), n = 265 . | P . |
|---|---|---|---|---|---|
| t(9;11) | 55 (11) | – | – | – | † |
| t(v;11)(v;q23)* | 52 (10) | 6 (11) | 2 (1) | 1 | < .0001 |
| t(6;9)* | 16 (3) | 3 (6) | – | – | † |
| inv(3) or t(3;3) | 11 (2) | 34 (63) | – | – | † |
| −5 or 5q− | 8 (2) | 5 (9) | 53 (22) | 169 (64) | < .0001 |
| −7 | 36 (67) | 42 (18) | 106 (40) | < .0001 | |
| 7q− | 14 (3) | 5 (9) | 40 (17) | 49 (18) | < .0001 |
| +8 or +8q | 157 (31) | 3 (6) | 48 (20) | 42 (16) | < .0001 |
| +11 or +11q | 23 (5) | 0 | 14 (6) | 20 (8) | < .0001 |
| abnl(12p) | 18 (4) | 4 (7) | 27 (11) | 73 (28) | < .0001 |
| +13 or +13q | 21 (4) | 0 | 12 (5) | 10 (4) | .44 |
| abnl(17p) | 6 (1) | 6 (11) | 12 (5) | 124 (47) | < .0001 |
| −18 or 18q− | 1 (0.2) | 5 (9) | 2 (1) | 56 (21) | < .0001 |
| −20 or 20q− | 23 (5) | 4 (7) | 9 (4) | 57 (22) | < .0001 |
| +21 or +21q | 28 (6) | 0 | 21 (9) | 23 (9) | < .0001 |
| +22 or +22q | 4 (1) | 0 | 14 (6) | 13 (5) | < .0001 |
AML indicates acute myeloid leukemia; MDS, myelodysplasia-related cytogenetic abnormalities according to Swerdlow et al3 ; MK, monosomal karyotype; WHO, World Health Organization; and –, not applicable. Percentages may not add to 100 because of rounding.
One case exhibited a t(6;9) and t(v;11)(v;q23) concurrently.
Response to induction therapy
Before start of intensive induction therapy, 11 patients died. Response to induction therapy for MK+ and MK− was as follows: CR, 33% (102 of 314) and 58% (427 of 733); refractory disease (RD), 52% (164 of 314) and 33% (244 of 733); early/hypoplastic death (ED/HD), 15% (48 of 314) and 9% (62 of 733), respectively. In uni- as well as multivariable analysis, MK+ had a significant impact on achievement of CR (P < .0001, univariable; P = .007, multivariable, Table 5). The number of AM did not impact on response to induction therapy among MK+ patients (in patients with one and 2 AM CR rate 34% each; in patients with 3 or more AM CR rate 30%; P = .76).
Multivariable logistic regression analysis on response to induction therapy
| Factor . | Odds ratio . | 95% CI . | P . |
|---|---|---|---|
| s-/t-AML | 0.65 | 0.46-0.92 | .02 |
| Age, difference of 10 y | 0.67 | 0.60-0.74 | < .0001 |
| Log10 of WBC | 0.75 | 0.61-0.91 | .007 |
| Log10 of platelets | 1.18 | 0.84-1.61 | .35 |
| Monosomal karyotype* | 0.58 | 0.39-0.86 | .007 |
| Complex karyotype† | 0.64 | 0.43-0.94 | .02 |
| t(6;9) | 1.27 | 0.45-3.62 | .65 |
| t(9;11) | 1.67 | 0.86-3.25 | .13 |
| t(v;11)(v;q23) | 0.81 | 0.45-1.45 | .47 |
| inv(3) or t(3;3) | 0.16 | 0.06-0.39 | .0001 |
| NPM1 mutation | 2.73 | 1.42-5.27 | .003 |
| FLT3-ITD | 0.69 | 0.43-1.10 | .12 |
| Factor . | Odds ratio . | 95% CI . | P . |
|---|---|---|---|
| s-/t-AML | 0.65 | 0.46-0.92 | .02 |
| Age, difference of 10 y | 0.67 | 0.60-0.74 | < .0001 |
| Log10 of WBC | 0.75 | 0.61-0.91 | .007 |
| Log10 of platelets | 1.18 | 0.84-1.61 | .35 |
| Monosomal karyotype* | 0.58 | 0.39-0.86 | .007 |
| Complex karyotype† | 0.64 | 0.43-0.94 | .02 |
| t(6;9) | 1.27 | 0.45-3.62 | .65 |
| t(9;11) | 1.67 | 0.86-3.25 | .13 |
| t(v;11)(v;q23) | 0.81 | 0.45-1.45 | .47 |
| inv(3) or t(3;3) | 0.16 | 0.06-0.39 | .0001 |
| NPM1 mutation | 2.73 | 1.42-5.27 | .003 |
| FLT3-ITD | 0.69 | 0.43-1.10 | .12 |
Furthermore, we evaluated the impact of MK on response in patients with CK and in patients with MDS-related cytogenetic abnormalities. CK+MK+ patients had an in trend inferior CR rate compared with CK+MK− patients (33% vs 45%; P = .056); in contrast, among MK+ patients the presence of a CK did not impact the CR rate (33% for MK+CK+ vs 30% for MK+CK−; P = .68). Similarly, in patients with MDS-related cytogenetic abnormalities MK+ patients had a significantly inferior CR rate compared with MK− patients (34% for MDS-related MK+ vs 49% MDS-related MK−; P = .007); the presence of MDS-related cytogenetic abnormalities in MK+ AML did not impact the CR rate (34% MK+ MDS-related vs 25% MK+ not MDS-related; P = .20).
With respect to the MK-R subgroup analysis within the subset of all patients without recurrent genetic abnormalities according to WHO classification and available response data (n = 811), response to induction was as follows: CR, 35% (90 of 255) and 54% (298 of 556), P < .0001; RD, 49% (126 of 255) and 37% (204 of 556), P = .0007; ED/HD, 15% (39 of 255) and 10% (54 of 556), P = .024, for MK-R+ and MK-R− patients, respectively. Of note, in a multivariable logistic regression model, MK-R did only in trend impact on achievement of CR (P = .07) with an odds ratio of 0.68 (95% CI, 0.44-1.04).
In the MK− group, the unfavorable parameters adverse cytogenetics (P = .0004) and CK (P = .01) retained their negative prognostic impact on achievement of CR.
Survival analysis
The median follow-up for survival in the subgroup of 1058 cytogenetically abnormal patients was 4.34 years (95% CI, 4.07 to 4.67 years); the estimated 4-year RFS and OS rates were 28% (95% CI, 24%-32%) and 23% (95% CI, 21%-26%).
The outcome of MK+ patients was significantly inferior: the 4-year RFS was 18% (95% CI, 12%-28%) and 30% (95% CI, 26%-35%; P = .0007), and the 4-year OS was 9% (95% CI, 7%-13%) and 29% (95% CI, 26%-33%; P < .0001) for MK+ and MK− patients, respectively. Among MK+ patients, those with 3 or more AM had an inferior outcome compared with those with only one or 2 AM (4-year OS of 4% [95% CI, 1%-10%] and 13% [95% CI, 9%-19%], respectively; P = .002).
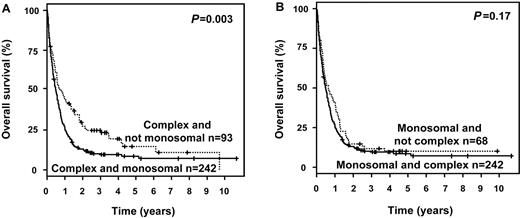
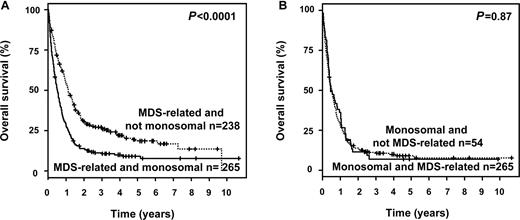
Furthermore, we analyzed the impact of MK status in patients with CK and in patients with MDS-related cytogenetic abnormalities. MK maintained its negative prognostic impact on OS in CK patients (P = .003; Figure 1A), and in those exhibiting MDS-related cytogenetic abnormalities (P < .0001; Figure 2A). In contrast, in MK+ patients neither CK (P = .17; Figure 1B) nor MDS-related cytogenetic abnormalities (P = .87; Figure 2B) impacted OS.
Survival curves according to MK and CK. (A) Impact of MK in patients exhibiting a CK and (B) impact of CK in patients exhibiting a MK.
Survival curves according to MK and CK. (A) Impact of MK in patients exhibiting a CK and (B) impact of CK in patients exhibiting a MK.
Survival curves according to MK and WHO category MDS-related changes. (A) Impact of MK in patients exhibiting MDS-related cytogenetic abnormalities and (B) impact of MDS-related cytogenetic abnormalities in patients exhibiting a MK.
Survival curves according to MK and WHO category MDS-related changes. (A) Impact of MK in patients exhibiting MDS-related cytogenetic abnormalities and (B) impact of MDS-related cytogenetic abnormalities in patients exhibiting a MK.
In the MK− group, the unfavorable parameters adverse cytogenetics (P < .0001) and CK (P = .0002) retained their negative prognostic impact on OS.
In multivariable Cox regression analysis, MK+ was an independent adverse prognostic factor for OS (hazard ratio [HR] 1.60, 95% CI, 1.30-1.97; P < .0001; Table 6 for the multivariable model). The same effect was found when using the variable “MK-R” instead of “MK” (HR 1.53, 95% CI, 1.23-1.92; P = .0002).
Multivariable Cox model for overall survival (stratified by trials for younger vs elderly patients)
| Factor . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| s-/t-AML | 1.28 | 1.07-1.53 | .007 |
| Age, difference of 10 y | 1.20 | 1.10-1.31 | < .0001 |
| Log10 of WBC | 1.33 | 1.19-1.50 | < .0001 |
| Log10 of platelets | 0.81 | 0.69-0.87 | .02 |
| Monosomal karyotype* | 1.60 | 1.30-1.97 | < .0001 |
| Complex karyotype† | 1.57 | 1.27-1.94 | < .0001 |
| t(6;9) | 1.34 | 0.73-2.44 | .35 |
| t(9;11) | 0.79 | 0.51-1.23 | .29 |
| t(v;11)(v;q23) | 1.36 | 0.95-1.97 | .10 |
| inv(3) or t(3;3) | 1.99 | 1.34-2.96 | .0007 |
| NPM1 mutation | 0.59 | 0.41-0.85 | .006 |
| FLT3-ITD | 1.45 | 1.12-1.87 | .004 |
| Factor . | Hazard ratio . | 95% CI . | P . |
|---|---|---|---|
| s-/t-AML | 1.28 | 1.07-1.53 | .007 |
| Age, difference of 10 y | 1.20 | 1.10-1.31 | < .0001 |
| Log10 of WBC | 1.33 | 1.19-1.50 | < .0001 |
| Log10 of platelets | 0.81 | 0.69-0.87 | .02 |
| Monosomal karyotype* | 1.60 | 1.30-1.97 | < .0001 |
| Complex karyotype† | 1.57 | 1.27-1.94 | < .0001 |
| t(6;9) | 1.34 | 0.73-2.44 | .35 |
| t(9;11) | 0.79 | 0.51-1.23 | .29 |
| t(v;11)(v;q23) | 1.36 | 0.95-1.97 | .10 |
| inv(3) or t(3;3) | 1.99 | 1.34-2.96 | .0007 |
| NPM1 mutation | 0.59 | 0.41-0.85 | .006 |
| FLT3-ITD | 1.45 | 1.12-1.87 | .004 |
Evaluation of allogeneic HSCT in younger MK+ patients
Outcome analyses according to allogeneic HSCT were restricted to patients between 18 and 60 years. For further analyses, patients with ED/HD during induction therapy (n = 27) were excluded. In all treatment protocols allogeneic HSCT was intended in younger AML patients with adverse-risk cytogenetics. Allogeneic HSCT was performed in 103 (61%) of 168 MK+ patients (31 from matched related [MRD], 70 from matched unrelated donors [MUD], 2 from haploidentical sibling donors [HAPLO]; Table 7). Of 82 patients achieving a CR after induction therapy, 57 proceeded to allogeneic HSCT (MRD, n = 16; MUD, n = 40; HAPLO, n = 1). Of 86 patients with refractory disease after induction therapy, 46 proceeded to allogeneic HSCT (MRD, n = 15; MUD, n = 30; HAPLO, n = 1). Patients receiving a HAPLO transplantation (n = 2) were included for further analyses into the group of patients receiving a MUD transplantation (n = 70). The median time interval between diagnosis and allogeneic HSCT was 115 days for MUD and 99 days for MRD transplantation. Age differed significantly among treatment strategies (P = .002): patients not proceeding to allogeneic HSCT had a median age of 55 years, whereas patients receiving an allogeneic HSCT had a median age of 50 years. However, there was no association between age and source of donor (median age: MRD, 49 years; MUD, 51 years; P = .22).
Selected characteristics in younger (age < 61 years) AML patients with MK according to treatment strategy (n = 163)
| Characteristics . | Allogeneic HSCT from MRD, n (%) . | Allogeneic HSCT from MUD, n (%) . | No HSCT, n (%) . | P . |
|---|---|---|---|---|
| Sex | ||||
| Male | 17 (55) | 41 (57) | 37 (57) | .99 |
| Female | 14 (45) | 31 (43) | 28 (43) | |
| Median age, y (range) | 49 (19-59) | 51 (23-61) | 55 (27-61) | .002 |
| WBC | .33 | |||
| Median (range) | 6.2 (1.8-108) | 8.0 (0.9-210) | 5.0 (0.5-533) | |
| No. missing | 0 | 0 | 0 | |
| Percentage of PB blasts | .51 | |||
| Median (range) | 24 (0-91) | 35 (0-99) | 34 (0-94) | |
| No. missing | 4 | 2 | 4 | |
| Percentage of BM blasts | .69 | |||
| Median (range) | 61 (21-100) | 65 (8-99) | 68 (15-100) | |
| No. missing | 2 | 7 | 3 | |
| Type of AML | .08 | |||
| De novo | 20 (65) | 60 (83) | 54 (83) | |
| s-/t-AML | 11 (35) | 12 (17) | 11 (17) | |
| Complex karyotype* | 18 (58) | 53 (74) | 50 (77) | .15 |
| MDS-related cytogenetic abnormalities† | 24 (77) | 56 (78) | 52 (80) | .94 |
| Response to induction | 16 (52) | 41 (57) | 25 (38) | .09 |
| Characteristics . | Allogeneic HSCT from MRD, n (%) . | Allogeneic HSCT from MUD, n (%) . | No HSCT, n (%) . | P . |
|---|---|---|---|---|
| Sex | ||||
| Male | 17 (55) | 41 (57) | 37 (57) | .99 |
| Female | 14 (45) | 31 (43) | 28 (43) | |
| Median age, y (range) | 49 (19-59) | 51 (23-61) | 55 (27-61) | .002 |
| WBC | .33 | |||
| Median (range) | 6.2 (1.8-108) | 8.0 (0.9-210) | 5.0 (0.5-533) | |
| No. missing | 0 | 0 | 0 | |
| Percentage of PB blasts | .51 | |||
| Median (range) | 24 (0-91) | 35 (0-99) | 34 (0-94) | |
| No. missing | 4 | 2 | 4 | |
| Percentage of BM blasts | .69 | |||
| Median (range) | 61 (21-100) | 65 (8-99) | 68 (15-100) | |
| No. missing | 2 | 7 | 3 | |
| Type of AML | .08 | |||
| De novo | 20 (65) | 60 (83) | 54 (83) | |
| s-/t-AML | 11 (35) | 12 (17) | 11 (17) | |
| Complex karyotype* | 18 (58) | 53 (74) | 50 (77) | .15 |
| MDS-related cytogenetic abnormalities† | 24 (77) | 56 (78) | 52 (80) | .94 |
| Response to induction | 16 (52) | 41 (57) | 25 (38) | .09 |
AML indicates acute myeloid leukemia; MDS, myelodysplastic syndrome; MRD, matched related donor; MUD, matched unrelated donor; PB, peripheral blood; s-AML, secondary AML; t-AML, therapy-related AML; and WBC, white blood count. Percentages may not add to 100 because of rounding.
According to Döhner et al.6
According to Swerdlow et al.3
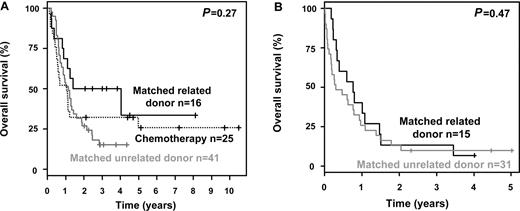
The 4-year OS rates were 13% (95% CI, 7%-24%) for patients not proceeding to allogeneic HSCT (n = 65) measured from the date of diagnosis and 15% (95% CI, 8%-25%) for patients receiving an allogeneic HSCT (n = 103), measured from the date of transplantation. There was a marked difference in survival between patients achieving a CR after induction therapy and those who did not (P < .0001). To account for the time dependency of allogeneic HSCT, we performed univariable Mantel-Byar analyses.18 Overall, allogeneic HSCT significantly improved OS (P = .0009). However, this beneficial impact was not equally distributed in patients achieving a CR or not after induction therapy. In patients achieving a CR after induction therapy, no beneficial impact of allogeneic HSCT could be demonstrated (P = .85). There was a trend toward a better survival measured from date of transplantation in patients receiving an allogeneic HSCT from MRD compared with MUD (P = .098). The 4-year survival rates for patients achieving a CR after induction therapy were 50% after allogeneic HSCT from MRD (95% CI, 31%-82%), 15% from MUD (95% CI, 7%-32%), and 32% (95% CI, 18%-57%) for those patients not proceeding to allogeneic HSCT, respectively (Figure 3A).
Survival curves according to induction result and post remission treatment. (A) OS measured from date of first CR in younger MK patients achieving a CR after induction therapy according to treatment strategy. (B) OS measured from date of transplantation in younger MK patients refractory to induction therapy according to type of donor.
Survival curves according to induction result and post remission treatment. (A) OS measured from date of first CR in younger MK patients achieving a CR after induction therapy according to treatment strategy. (B) OS measured from date of transplantation in younger MK patients refractory to induction therapy according to type of donor.
Patients not achieving a CR after induction therapy had a dismal outcome. Median survival of those patients not proceeding to allogeneic HSCT was 3.5 months measured from diagnosis and all patients (n = 40) died within 2 years. Although not in CR, 46 (53%) of the 86 patients proceeded to allogeneic HSCT and median survival was 9.1 months for MRD and 3.7 months for MUD transplants measured from the data of allogeneic HSCT, respectively. In this subset, Mantel-Byar analysis revealed a beneficial effect of allogeneic HSCT on OS (P < .0001). However, after 2 years, only 2 and 3 patients were alive after transplantation from MRD and MUD, respectively (Figure 3B). A multivariable model on OS in all patients who actually received an allogeneic HSCT revealed achievement of CR after induction therapy (HR, 0.48; P = .001) as a favorable and allogeneic HSCT from MUD (HR, 1.70; P = .05) as an unfavorable factor, whereas type of AML, age, presence of CK, white blood count, and platelets at diagnosis had no significant impact. Cumulative incidences of relapse (CIR) 2 years after allogeneic HSCT were 52% and 61% (P = .37), and those of death (CID) 15% and 20% (P = .53) for MRD and MUD, respectively.
Discussion
Recently, the new cytogenetic category “MK” was reported and shown to be associated with a dismal prognosis.7 Our data largely confirm those from the pivotal study, but also extend on these findings in particular in the light of the new WHO classification and with respect to the impact of allogeneic HSCT.
In our study, MK was identified in approximately one-third of cytogenetically abnormal AML patients, excluding those with CBF-AML, APL and isolated losses of sex chromosomes. MK was significantly associated with high-risk abnormalities, such as −5 or 5q−, −7, abnl(17p), and CK, consistent with previous reports.7,22 There was also a significant association of MK with MDS-related chromosome abnormalities, and, among the subgroup of AML with recurrent genetic abnormalities, with inv(3) or t(3;3) that in approximately two-thirds of cases exhibit −7 and thus fulfill the MK criteria.23 Virtually all MK+ cases (98%) were contained within the cytogenetic adverse-risk group as defined by the European LeukemiaNet criteria.6 For a large proportion of our cases, data on the mutational status of the NPM1 and FLT3 genes were available. Of note, NPM1 mutations and FLT3-ITD were found at very low frequencies in MK+ AML, highly significantly lower compared with MK− AML. Thus, the spectrum of both cytogenetic and molecular genetic changes reflects another disease biology in MK+ AML.
Regarding clinical parameters, MK was associated with higher age at diagnosis consistent with previous studies.7,22 Furthermore, MK+ patients had lower hemoglobin levels, lower median white blood counts, and lower percentages of blasts in peripheral blood as well as in BM.
As described previously7,22 MK status was an independent adverse prognostic factor and added prognostic information even in the subgroup of CK+ patients. Furthermore, we demonstrate that MK is an independent adverse prognostic factor in the subgroup of patients with MDS-related cytogenetic abnormalities. Thus, MK appears to outperform the categories “CK” and “MDS-related cytogenetic abnormalities” with respect to prognostication. MK remained a prognostic factor for OS but not for achievement of CR after induction therapy in a revised version (MK-R) when cases with recurrent genetic abnormalities [mainly AML with inv(3) or t(3;3)], and those with derivative chromosomes not leading to true monosomies were excluded. However, the magnitude of the unfavorable impact on survival was weaker for MK-R compared with MK, mainly because of the exclusion of the very unfavorable entity “recurrent genetic abnormality with inv(3) or t(3;3)” according to the WHO classification.3 In addition, an higher amount of AM impacted on survival within the MK+ group, thus expanding the findings reported by Breems et al.7
With respect to the question of including the MK definition into the risk categorization in AML, results in the MK− group are of special interest. In our analyses, CK as well as the risk category “adverse” according to the ELN criteria6 retained their prognostic impact with respect to induction success and OS even in the MK− group. For CK, this is in contrast to the data reported by Breems et al.7 This difference may be due to a higher statistical power in our study based on a sample size almost twice as high as that reported by Breems et al.7 Therefore, the category CK may not be simply replaceable by the category MK for the adverse-risk definition.
To evaluate whether the adverse outcome of MK+ patients can be overcome by allogeneic HSCT, we performed survival analyses in younger MK+ patients. Of note, although CR rate in MK+ patients was only 32%, more than half of the patients proceeded to allogeneic HSCT. In our study only a limited beneficial effect of allogeneic HSCT in MK+ patients could be demonstrated. Of note, patients with refractory AML after induction therapy had a significant benefit from allogeneic HSCT with few long-term survivors. In contrast, we were not able to show a significant benefit for allogeneic HSCT in patients achieving a CR after induction therapy. This was mainly because of an inferior outcome of patients after allogeneic HSCT from MUD compared with that from MRD which was evident in uni- and multivariable analyses. This was in contrast to the encouraging data reported by Fang et al showing in a retrospective subgroup analysis a favorable outcome, especially for patients who received transplants in first CR.24 These conflicting data on the value of allogeneic HSCT in AML with MK were based on the one hand on a retrospective case series24 and on the other hand on our meta-analyses of patients taken into prospective clinical multicenter trials indicate the need for a prospective evaluation of this treatment strategy. Consistent with the report from Oran et al,25 we observed high CIR rates after allogeneic HSCT from MRD and MUD in our MK+ patients, indicating the urgent need for alternative treatment strategies to improve results. Although not statistically significant, the combination of higher CIR and CID rates in patients receiving an allogeneic HSCT from MUD compared with those from MRD might be a possible explanation for the inferior outcome after allogeneic HSCT from MUD.
In conclusion, our study confirms that MK is a strong independent adverse prognostic factor in adult AML. MK further dissects the subsets of AML with CK and with MDS-related cytogenetic abnormalities. Our study could demonstrate a marginal beneficial effect of allogeneic HSCT, therefore stressing the need for novel therapeutic strategies in this very poor prognostic group.
Presented in part at the 15th annual meeting of the European Hematology Association, Barcelona, Spain, June 11, 2010.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to all members of the German-Austrian AML Study Group (AMLSG) for providing leukemia specimens as well as clinical data, and they thank the patients for participating in the AMLSG trials.
This work was supported in part by grants 01GI9981 (Network of Competence Acute and Chronic Leukemias), 01KG0605 (IPD-Meta-Analysis: A model-based hierarchical prognostic system for adult patients with acute myeloid leukemia [AML]) from the Bundesministerium für Bildung und Forschung (BMBF), Germany, and grant DJCLS R 08/23v from the Deutsche José Carreras Leukämie-Stiftung.
Authorship
Contribution: S.K. collected, analyzed, and interpreted data, designed research, and wrote the manuscript; M.Z. analyzed and interpreted data and wrote the manuscript; K.D., J.K., C.-H.K., H.A.H., G.H., M.v.L.-T., S.W., M.R., U.G., K.G., D.N., and A.G. provided study materials or patients and collected data; B.S., G.G., D.S., C.M., and V.T. collected data; H.D. and R.F.S. provided study materials or patients, designed research, collected, analyzed, and interpreted data, and wrote the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of AMLSG institutions and investigators participating in this study appears in the supplemental Appendix.
Correspondence: Richard F. Schlenk, MD, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: richard.schlenk@uniklinik-ulm.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal