Sometimes referred to as Kostmann syndrome (after the Swedish pediatrician who described a family of individuals with neutropenia in 1956),2 SCN is one of the rare inherited bone marrow failure syndromes. Typically presenting within the first few months of life, babies present with a life-threatening infection of the skin or gastrointestinal or respiratory organs. Classically, there is profound neutropenia (< 200/μL) in the periphery and, in the bone marrow, there is a developmental arrest at the promyelocyte stage. Despite broad spectrum antibiotics, many died from infection. Treatment with recombinant human granulocyte colony stimulating factor (filgrastim) in the early 1990s changed the natural history of the disease, enabling children to live into adulthood. Shortly after the widespread adoption of filgrastim as treatment of choice, Dong and colleagues described somatic nonsense mutations in the GCSFR among patients who developed myelodysplastic syndromes (MDS) or frank AML.3 French and National Institutes of Health–based epidemiologic studies reported that an accumulated risk of developing MDS/AML was as high as 40%.4,5 Like the other inherited bone marrow failure syndromes (such as Fanconi anemia, dyskeratosis congenita, and Shwachman-Diamond syndrome), SCN constitutes a cancer predisposition syndrome.
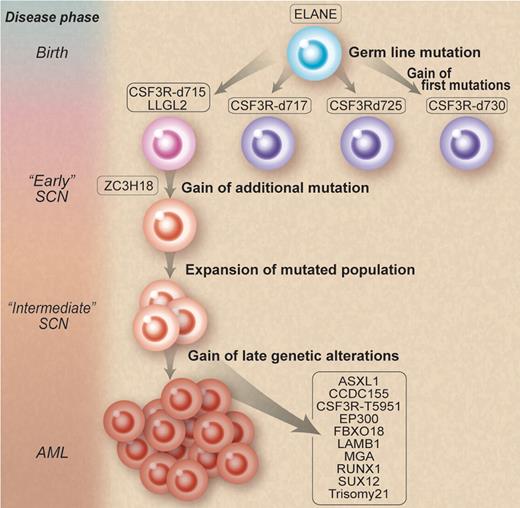
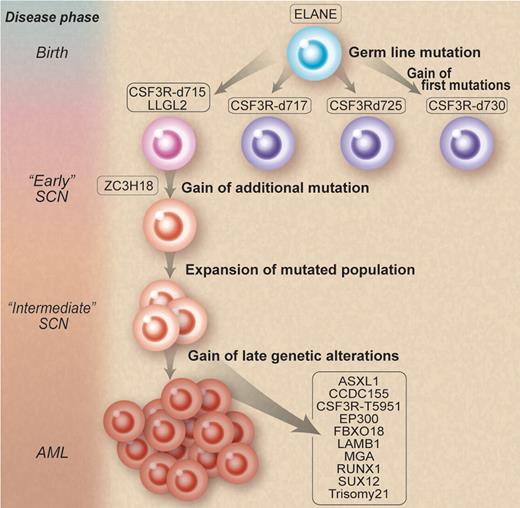
How does one go from famine (SCN) to feast (AML)? Working backward, Beekman et al isolated 12 somatic nonsynonymous mutations in the leukemic blasts of a patient. They then retrieved cryopreserved bone marrow specimens from 9 and 15 years before the development of AML. They found that 3 of these mutations, GCSFRd715, LLGL2, and ZC3H18, were present in low frequencies and coexisted in the same myeloid progenitors. They further demonstrated an increase in these clones over time from 15 to 9 years before the onset of AML, thereby supporting the notion of a selective outgrowth of clones harboring these 3 mutations. They also identified small amounts of other GCSFR mutations at 15 and 9 years before leukemia. Most fascinating of all is that only the GCSFRd715 persisted clonally. This demonstration provides supportive evidence for competitive outgrowth of the other mutants and that, true to evolution, some survive and others die off (see figure). Furthermore, they discovered a second “hit” to the GCSFRd715 involving a novel mutation (T595I), only present at the time of AML.
Natural selection of clones. Mutations involving granulocyte colony-stimulating factor receptor (GCSFR) emerge during the course of severe congenital neutropenia (SCN) treated by recombinant human G-CSF. However, some mutations thrive and persist, while others die off. Professional illustration by Debra T. Dartez adapted from Figure 4 in Beekman et al, which begins on page 5071.
Natural selection of clones. Mutations involving granulocyte colony-stimulating factor receptor (GCSFR) emerge during the course of severe congenital neutropenia (SCN) treated by recombinant human G-CSF. However, some mutations thrive and persist, while others die off. Professional illustration by Debra T. Dartez adapted from Figure 4 in Beekman et al, which begins on page 5071.
Like Good's immunodeficient children whom he called “experiments of nature,”6 key insights in human biology emerge from molecular and cellular study of a single patient. While multistep clonal pathogenesis of a cancer has long been suspected since Vogelstein's classic study of colorectal tumors,7 the facility of sampling hematopoietic clonal descendants coupled to new advances in exome or whole genome sequencing has now provided the most compelling evidence. Comparable work recently reported by Walter et al demonstrated that recurrent mutations existed in founding and daughter clones from patients with MDS that progressed to AML.8
Touw's group has continued to reveal the complex role played by the GCSFR in the pathogenesis of SCN. While this group sought to recapitulate the transformation with a knock-in of the mutated GCSFRd715, the mice did not develop MDS/AML even after superpharmacologic doses of GCSF was administered.9 A similar disappointment occurred when Link's laboratory created mice with the knock-in of a mutated form of neutrophil-expressed elastase (ELANE), commonly affected in both SCN and cyclic neutropenia.10 The mice did not develop neutropenia. One important lesson from these studies is that murine myelopoiesis and pathology do not provide a precise model for human myelopoiesis and disease. Clinical investigation trumps, even when it is on a single patient.
Several unanswered questions remain in the famine-to-feast story of SCN. (1) Which came first: the sick stem cell11 or the superpharmacologic dose of filgrastim that promoted genetic instability? The connection between dose of filgrastim and risk of MDS/AML was suggested by the French and American studies.4,5 Indeed, the patient described here required a higher-than-average dose. (2) What is the initiating genetic lesion of SCN? Mutations in multiple genes have been identified in patients with SCN. The genes encode a protease (ELANE), a mitochondrial protein (Hax-1), a transcription factor (Gfi-1), a metabolic enzyme (G6PC), a cytoskeletal protein (WASP), and the GCSFR. No bone marrow samples from the time of diagnosis or at the start of filgrastim therapy were available to be interrogated. Thus, we cannot be certain that mutations of GCSFR, LLGL2, or ZC3H18 were not present ab initio. It is possible that they or some other agent, including epigenetic silencing or aberrant expression of a microRNA, constitutes a primordial risk factor. (3) Why do MDS/AML not arise in pure cyclic neutropenia as they do in SCN, even though both may involve ELANE mutations? (4) How does GCSFR T595I, occurring in the external domain of the receptor, perturb GCSFR signaling? And how does this mutation affect the signaling effects of GCSFRd715? (5) Especially for the clinician: what is the cost-benefit of filgrastim? Does the unknown mechanism of action for filgrastim's ability to restore neutrophil production also contribute to myeloid progenitor cell disease?
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■