Severe congenital neutropenia (SCN) is a BM failure syndrome with a high risk of progression to acute myeloid leukemia (AML). The underlying genetic changes involved in SCN evolution to AML are largely unknown. We obtained serial hematopoietic samples from an SCN patient who developed AML 17 years after the initiation of G-CSF treatment. Next- generation sequencing was performed to identify mutations during disease progression. In the AML phase, we found 12 acquired nonsynonymous mutations. Three of these, in CSF3R, LLGL2, and ZC3H18, co-occurred in a subpopulation of progenitor cells already in the early SCN phase. This population expanded over time, whereas clones harboring only CSF3R mutations disappeared from the BM. The other 9 mutations were only apparent in the AML cells and affected known AML-associated genes (RUNX1 and ASXL1) and chromatin remodelers (SUZ12 and EP300). In addition, a novel CSF3R mutation that conferred autonomous proliferation to myeloid progenitors was found. We conclude that progression from SCN to AML is a multistep process, with distinct mutations arising early during the SCN phase and others later in AML development. The sequential gain of 2 CSF3R mutations implicates abnormal G-CSF signaling as a driver of leukemic transformation in this case of SCN.
Introduction
Severe congenital neutropenia (SCN) is a BM failure syndrome characterized by strongly reduced neutrophil counts and recurrent, potentially life-threatening, opportunistic bacterial infections. Treatment with G-CSF elevates peripheral neutrophil counts and reduces the risk of infections.1 Leukemic progression of SCN is a major concern, with an estimated overall cumulative incidence of approximately 20% after 15 years of G-CSF treatment.2
Constitutional mutations in the gene encoding neutrophil elastase (ELANE) are common defects in SCN.3 In addition, the acquisition of nonsense mutations in the gene encoding the G-CSF receptor (CSF3R) is a unique feature in SCN patients.4,,–7 These mutations lead to the expression of truncated CSF3R proteins, also known as the δ forms. In cell-line models, truncated CSF3R proteins are hampered in transducing the signals required for proper neutrophil differentiation, confer increased proliferative responses to G-CSF treatment, but do not cause leukemia in mice.4,–6,8,,–11 CSF3Rδ mutations can be detected in approximately 30% of SCN patients. In some cases, distinct clones with different CSF3Rδ mutations are present for many years.7,12 After evolution of SCN toward acute myeloid leukemia (AML), CSF3Rδ mutations are found in approximately 80% of patients.12 Until now, all reported SCN/AML patients harboring a CSF3Rδ mutation in the SCN phase also carry this mutation in the leukemic phase. These observations suggest that leukemic progression in SCN follows a unique pattern, with CSF3Rδ mutations as an early event, followed by additional genetic and epigenetic events that are essential for full leukemic transformation. Chromosomal aberrations such as loss of chromosome 7 and gain of chromosome 21 are apparent in AML arising from SCN and other BM failure syndromes such as Fanconi anemia and Shwachman-Diamond syndrome.13 However, mutations that are quite commonly seen in de novo AML have not been reported in AML arising from SCN.14 Therefore, the additional molecular events involved in leukemic progression of SCN remain largely unknown.
To identify the sequential genetic events in leukemic progression of SCN to AML, we collected serial hematopoietic samples from an SCN patient who developed AML after 17 years of G-CSF therapy. Using whole-exome sequencing (WES), we found 12 somatic nonsynonymous mutations in the leukemic blasts of this patient. Three of these mutations, the known CSF3R mutation and mutations in LLGL2 and ZC3H18, were already present at low frequencies in the early SCN phase 15 years before AML was diagnosed. Myeloid colony analysis showed that these 3 “early” mutations coexisted in the same hematopoietic progenitors in a small subpopulation of BM cells. Six years later, in the “intermediate” SCN phase, still 9 years before the AML became overt, we observed an expansion of the clone harboring all 3 mutations. The other 9 mutations were only apparent in the AML cells. The latter “late-appearing” mutations comprise a second, novel CSF3R mutation in addition to a series of new and known AML-associated mutations. The novel CSF3R mutation is located on the already mutated CSF3R-d715 allele and causes growth factor independence of myeloid progenitors.
Methods
Case reports
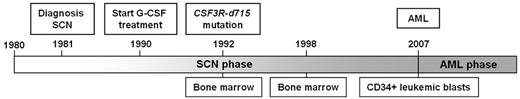
A 27-year-old male SCN patient was diagnosed with AML 17 years after the start of G-CSF treatment (10 μg/kg/d), on which he reached normal neutrophil counts. The patient had a constitutional heterozygous ELANE mutation, G174R. At the age of 12, 2 years after G-CSF treatment was initiated, a CSF3R δ mutation (CSF3R-d715) was discovered in the BM.6 At the time of AML diagnosis, the peripheral blood contained 24% blasts and dysplasia was observed in the BM. G-CSF treatment was stopped at this point. Six weeks later, a BM analysis revealed 17% blasts. Immunophenotypically, these blasts were of a myeloid nature (ie, positive for CD34, CD117, CD13, CD133, CD33, MPO, and CD90). Because no HLA-identical donor was available, the patient received a matched unrelated donor allogeneic BM transplantation. Induction therapy was given according to the induction therapy scheme HOVON42A of the Hemato-Oncology Foundation for Adults in the Netherlands.15 At initiation of induction therapy, the BM contained 15.7% blasts with 10%-50% dysplasia in all lineages. Routine cytogenetic and molecular diagnostics revealed a trisomy 21 (47, XY, +21[14]/46, XY[4]) with no additional abnormalities (AML-ETO, CBFB/MYH11, FLT3ITD, FLT3TKD, mutations in NPM1, NRAS, KRAS, c-KIT, JAK2, and CEBPA). After the second induction cycle, trisomy 21 was undetectable in a BM cytogenetic analysis. The matched unrelated donor transplantation was administered after myeloablative conditioning with chemotherapy and total body irradiation. Two months after the transplantation, 28% blasts were detected in the BM, indicating a recurrence of the AML, and the patient died 3.5 months after the transplantation. Figure 1 gives a schematic overview of the disease history.
Chronologic overview of the clinical course of a SCN/AML patient. Distinct events in the disease course are indicated above the timeline, such as the diagnosis of SCN, the initiation of G-CSF therapy, the discovery of the CSF3R-d715 mutation, and the diagnosis of AML.
Chronologic overview of the clinical course of a SCN/AML patient. Distinct events in the disease course are indicated above the timeline, such as the diagnosis of SCN, the initiation of G-CSF therapy, the discovery of the CSF3R-d715 mutation, and the diagnosis of AML.
Patient cell samples
Ficoll gradient–separated BM cells from the SCN phases and CD34+ leukemic blasts from the peripheral blood in the leukemic phase were used. Control DNA was isolated from BM-derived fibroblasts. All cell samples were obtained and frozen according to established procedures for viable cell cryopreservation, as described previously.16 The study was performed under the permission of the Institutional Review Board of the Erasmus Medical Center, registration number MEC-2008-387 for biobanking and MEC-2012-030 for the genetic analysis of leukemic progression in SCN patients.
Nucleotide sequencing
WES.
Sequencing libraries were prepared according to the SureSelect Target Enrichment system (protocol Version 2.2.1, November 2010; Illumina). In brief, 3 μg of genomic DNA was sheared to fragments of approximately 170 bp using the S-series Single Tube Sample Preparation System, Model S2 (Covaris). Fragment sizes were checked on the Bioanalyzer (Agilent Technologies). Adapter-ligated libraries were prepared according to the manufacturer's protocol using the Paired-End Genomic DNA Sample Prep Kit PE-102-1001 (Illumina); 5 cycles of amplification were used. Five hundred nanograms of prepped library was taken for hybridization with the SureSelect Human All Exon Kit (G3362A; Agilent Technologies). A 5.5pM sample concentration was loaded for sequencing on the Hiseq2000 (Illumina) using 101-bp paired-end reads.
Sequencing reads were processed with CASAVA Pipeline (Version 1.7; Illumina). For alignment, the Hg18/NCBI36 assembly (March 2006) was used. Detection of single nucleotide variants, deletions, and insertions was performed with otherwise default settings while snpCovCutoff and indelsCovCutoff were switched off. Variations detected in the AML sample in 2 independent sequence runs were further analyzed after removal of germline variations (present in the fibroblasts) and single nucleotide polymorphisms17 and then nonsynonymous variants were determined. The Integrative Genome Browser was used for sequence read visualization.18
Sanger sequencing.
WES results were validated by Sanger sequencing, which was performed according to the manufacturer's protocol (Applied Biosystems) using the primers indicated in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Before amplicon generation, genomic DNA or cDNA was first amplified using a Whole Genome Amplification kit (WGA2; Sigma-Aldrich). DNA was purified with a PCR purification kit (QIAGEN) according to the manufacturer's protocol and diluted to 50 ng/μL. One hundred nanograms of amplified DNA was used for amplicon generation. The cycling conditions were: 30 seconds at 95°C, 30 seconds at the indicated annealing temperature (supplemental Table 1), and 45 seconds at 72°C for 35 cycles. In some cases, the unamplified material was used directly for Sanger sequencing (supplemental Table 1).
Amplicon-based deep sequencing.
Amplicons were generated and purified according to the Amplicon Library Preparation Method Manual (May 2010 Version; Roche). Primers and annealing temperatures are indicated in supplemental Table 2; 35 cycles were used for amplification. DNA-enriched beads carrying the amplification products were generated according to the emPCR Amplification Method Manual Lib-A (May 2010 Version; Roche); a beads-to-amplicon ratio of 1:2 was used. Amplicons were analyzed with GS Junior (Roche). Sequence reads were analyzed using the GS Amplicon Variant Analyzer (Roche). For the SCN samples, coverage of at least 1600 was achieved to identify mutations present in minor clones within the BM. For the AML sample, coverage of 80 was considered sufficient to validate mutations.
Human myeloid colony assay
BM was thawed at 37°C, washed twice with IMDM (GIBCO Invitrogen) with 10% FCS (PAA Laboratories). For every 4 mL of culture medium, 2.9 mL of MethoCult (H4230; StemCell Technologies), 980 μL of IMDM, and human GM-CSF (Immunex), human G-CSF (Neupogen; Amgen), and human IL-3 (R&D Systems) in final concentrations of, respectively, 2 ng/mL, 200 ng/mL, and 25 ng/mL were used. Cells were plated at a density of 0.8 × 105/mL. After 2 weeks, genomic DNA of single colonies was isolated, followed by amplification using the Whole Genome Amplification kit and Sanger sequencing of CSF3R-d715, LLGL2, and ZC3H18, as described in “Sanger sequencing.” Results were validated in an independent round of whole genome amplification for: (1) colonies harboring a mutation, (2) colonies with unclear sequences, and (3) several randomly chosen nonmutated colonies to rule out amplification artifacts. All colonies harboring mutations in CSF3R, LLGL2, or ZC3H18 were also analyzed for the presence of the remaining 9 mutations found in the AML sample.
Murine colony assays
Four different CSF3R expression constructs (WT, d715, T595I, and d715-T595I) were generated and retrovirally transduced into BM cells of Csf3r-deficient FVB/N mice.19 Colony assays of these transduced progenitors were performed as described previously.20
Further details of these procedures are provided in supplemental Methods.
Results
WES reveals acquired mutations in SCN/AML
WES was performed on genomic DNA from the CD34+ leukemic blast fraction and the fibroblast control sample. Acquired nonsynonymous mutations were detected by identification of single nucleotide variants and small insertions and deletions, followed by subtraction of variants present in the control fibroblasts and known single nucleotide polymorphisms.17 Twelve nonsynonymous acquired mutations were identified and validated by Sanger sequencing (supplemental Figure 1 and Table 1). Except for the mutation in FBXO18, all mutations occurred in evolutionary conserved amino acids (supplemental Figure 2). With the exception of LAMB1, all mutant transcripts were detectably expressed in the leukemic blasts (supplemental Figure 3). Mutations in ASXL1 and RUNX1 are known in myeloid malignancies.21,22 Deletions in EP300, distinct from the 7-bp deletion found in this patient, have been reported in lymphomas.23,24 The ATT insertion in SUZ12 duplicates an isoleucine at amino acid position 597 located in the conserved VEFS-box. Mutations in this region, which is involved in the interaction between SUZ12 and the histone methyltransferase EZH2 in the polycomb repressor complex 2 (PRC2), have also been identified recently in myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) with 17q abnormalities.25 As expected, the previously identified CSF3Rδ mutation (CSF3R-d715) was present in the leukemic blasts but, remarkably a new CSF3R mutation, T595I, was now also present. Furthermore, the CSF3R-T595I mutation was located on the same allele as the δ mutation, as determined by Sanger sequencing of single amplicons generated from cDNA. Using exome sequencing data from 199 AML cases reported by The Cancer Genome Atlas (TCGA), a similar single CSF3R-T595I mutation was detected. In addition, mutations in ASXL1 (n = 5), CCDC155 (n = 1), LLGL2 (n = 1), MGA (n = 1), RUNX1 (n = 17), SUZ12 (n = 2), and ZC3H18 (n = 2) were found in the TCGA dataset (supplemental Table 3; R.G.V. and The Cancer Genome Atlas disease working group, unpublished data, January 2012.
Somatic nonsynonymous mutations in SCN/AML
| Gene symbol . | RefSeq reference transcript . | Genomic DNA change (NCBI36/hg18) . | cDNA change . | Mutation type . | Amino acid change . | Protein change . |
|---|---|---|---|---|---|---|
| ASXL1 | NM_015338.5 | g.chr20:30485948dupA | c.1772dupA | Indel frameshift | Frameshift and premature stop | p.Y591* |
| CCDC155 | NM_144688.4 | g.chr19:54601976C → T | c.820C → T | Missense | Arg → Trp | p.R274W |
| CSF3R-T595I | NM_000760.3 | g.chr1:36706021G → A | c.1853C → T | Missense | Thr → Ile | p.T595I† |
| CSF3R-d715 | NM_000760.3 | g.chr1:36704841G → A | c.2215C → T | Nonsense | Gln → * | p.Q716*† |
| EP300 | NM_001429.3 | g.chr22:39902447_39902453delTGGAGAC | c.5030_5036delTGGAGAC | Indel frameshift | Frameshift and premature stop | p.V1677Dfs*30 |
| FBXO18 | NM_032807.3 | g.chr10:6003435C → G | c.2372C → G | Missense | Ala → Gly | p.A791G |
| LAMB1 | NM_002291.2 | g.chr7:107387385delG | c.2445delC | Indel frameshift | Frameshift and premature stop | p.P815Pfs*65 |
| LLGL2 | NM_004524.2 | g.chr17:71070826G → C | c.665G → C | Missense | Arg → Pro | p.R222P |
| MGA | NM_001164273.1 | g.chr15:39787311C → T | c.2282C → T | Missense | Pro → Leu | p.P761L |
| RUNX1 | NM_001754.4 | g.chr21:35153662C → T | c.592G → A | Missense | Asp → Asn | p.D198N |
| SUZ12 | NM_015355.2 | g.chr17:27346889_27346891dupATT | c.1789_1791dupATT | Indel | Insertion Ile | p.597dupI |
| ZC3H18 | NM_144604.3 | g.chr16:87192175delC | c.777delC | Indel frameshift | Frameshift and premature stop | p.P259Pfs*15 |
| Gene symbol . | RefSeq reference transcript . | Genomic DNA change (NCBI36/hg18) . | cDNA change . | Mutation type . | Amino acid change . | Protein change . |
|---|---|---|---|---|---|---|
| ASXL1 | NM_015338.5 | g.chr20:30485948dupA | c.1772dupA | Indel frameshift | Frameshift and premature stop | p.Y591* |
| CCDC155 | NM_144688.4 | g.chr19:54601976C → T | c.820C → T | Missense | Arg → Trp | p.R274W |
| CSF3R-T595I | NM_000760.3 | g.chr1:36706021G → A | c.1853C → T | Missense | Thr → Ile | p.T595I† |
| CSF3R-d715 | NM_000760.3 | g.chr1:36704841G → A | c.2215C → T | Nonsense | Gln → * | p.Q716*† |
| EP300 | NM_001429.3 | g.chr22:39902447_39902453delTGGAGAC | c.5030_5036delTGGAGAC | Indel frameshift | Frameshift and premature stop | p.V1677Dfs*30 |
| FBXO18 | NM_032807.3 | g.chr10:6003435C → G | c.2372C → G | Missense | Ala → Gly | p.A791G |
| LAMB1 | NM_002291.2 | g.chr7:107387385delG | c.2445delC | Indel frameshift | Frameshift and premature stop | p.P815Pfs*65 |
| LLGL2 | NM_004524.2 | g.chr17:71070826G → C | c.665G → C | Missense | Arg → Pro | p.R222P |
| MGA | NM_001164273.1 | g.chr15:39787311C → T | c.2282C → T | Missense | Pro → Leu | p.P761L |
| RUNX1 | NM_001754.4 | g.chr21:35153662C → T | c.592G → A | Missense | Asp → Asn | p.D198N |
| SUZ12 | NM_015355.2 | g.chr17:27346889_27346891dupATT | c.1789_1791dupATT | Indel | Insertion Ile | p.597dupI |
| ZC3H18 | NM_144604.3 | g.chr16:87192175delC | c.777delC | Indel frameshift | Frameshift and premature stop | p.P259Pfs*15 |
All 12 somatic nonsynonymous mutations identified in the AML phase are listed. For each mutation, Refseq reference transcripts, the position of the mutation on genomic DNA, the cDNA and protein level, the mutation type, and the effect on the protein are indicated. See also supplemental Figures 1-3.
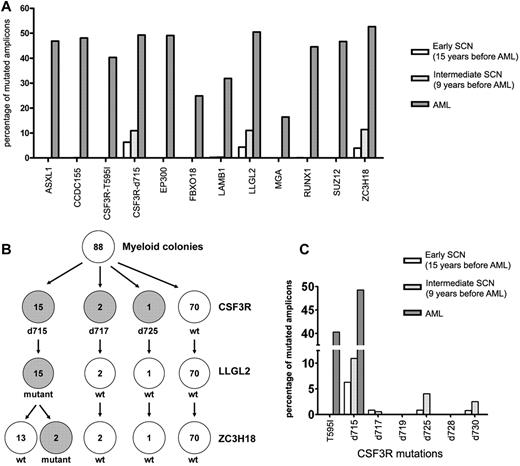
Amplicon-based sequencing reveals an early preleukemic clone that expands over time
Amplicon-based deep sequencing was applied to analyze the presence of all 12 somatic mutations in the BM samples obtained at 15 and 9 years before AML was diagnosed (Figure 1). Not only the known CSF3R-d715 mutation, but also mutations in LLGL2 and ZC3H18 were already present in these earlier disease phases (Figure 2A and supplemental Table 4). We investigated the clonal hierarchy of these mutations in single myeloid colonies cultured from the earliest BM sample (15 years before AML diagnosis). In the individual colonies (n = 88), the mutation status of CSF3R-d715, LLGL2, and ZC3H18 was determined. Fifteen colonies (17%) harbored both the CSF3R-d715 and the LLGL2 mutation, whereas none of the colonies exhibited exclusively either the LLGL2 or the CSF3R-d715 mutation (Figure 2B and supplemental Table 5). Two of the CSF3R-d715– and LLGL2-mutated colonies also carried the ZC3H18 mutation (Figure 2B and supplemental Table 5), indicating that this mutation had emerged later in time. None of the other 9 mutations found in the AML cells was apparent in these colonies (supplemental Table 5).
Acquisition of mutations in the evolution of SCN to AML. (A) The 12 somatic nonsynonymous mutations identified in the leukemic blasts were analyzed in the SCN phase using amplicon-based deep sequencing. The percentage of mutated amplicons per mutation is shown. Based on their frequencies in the AML population, all mutations are considered to be heterozygous, implying that the number of cells carrying the mutations is estimated to be twice the number of mutated amplicons. (B) Single myeloid colonies grown from the BM sample obtained 15 years before leukemia development were analyzed for the presence of mutations in CSF3R, LLGL2, and ZC3H18 (see supplemental Table 5 for more information). (C) The presence of different CSF3R mutations in the BM obtained 15 and 9 years before leukemia development and in the leukemic phase was investigated by amplicon-based deep sequencing. The percentage of mutated amplicons per mutation is shown. T595I indicates the CSF3R mutation T595I; d715-d730, CSF3Rδ mutations at amino acid positions 715-730.
Acquisition of mutations in the evolution of SCN to AML. (A) The 12 somatic nonsynonymous mutations identified in the leukemic blasts were analyzed in the SCN phase using amplicon-based deep sequencing. The percentage of mutated amplicons per mutation is shown. Based on their frequencies in the AML population, all mutations are considered to be heterozygous, implying that the number of cells carrying the mutations is estimated to be twice the number of mutated amplicons. (B) Single myeloid colonies grown from the BM sample obtained 15 years before leukemia development were analyzed for the presence of mutations in CSF3R, LLGL2, and ZC3H18 (see supplemental Table 5 for more information). (C) The presence of different CSF3R mutations in the BM obtained 15 and 9 years before leukemia development and in the leukemic phase was investigated by amplicon-based deep sequencing. The percentage of mutated amplicons per mutation is shown. T595I indicates the CSF3R mutation T595I; d715-d730, CSF3Rδ mutations at amino acid positions 715-730.
A previous study found that multiple CSF3Rδ mutations can be present in distinct progenitors in the BM of an individual SCN patient.7 In agreement with this finding, in the present study, we found myeloid colonies with CSF3R-d717 (n = 2) and CSF3R-d725 (n = 1; Figure 2B and supplemental Table 5). Each of these mutations and an additional δ mutation (CSF3R-d730) were detected in the SCN phase at low frequencies by amplicon-based deep sequencing (Figure 2C and supplemental Table 6). None of these variant CSF3R mutant clones harbored LLGL2 or ZC3H18 mutations, nor were they seen as dominant clones in AML (Figure 2 and supplemental Tables 5 and 6). No viably frozen cells were available from the BM sample obtained 9 years before AML development, and colony analysis could not be performed at this stage. However, by amplicon-based deep sequencing, we observed a parallel increase of the CSF3R-d715, LLGL2, and ZC3H18 mutations from 15 to 9 years before AML development (Figure 2A). Together with the finding that these mutations are present in the same myeloid progenitor cells (Figure 2B), this observation is consistent with a selective outgrowth of clones carrying these 3 mutations.
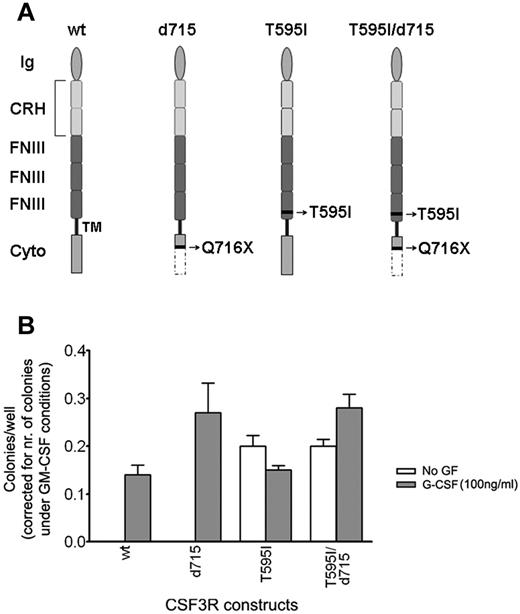
Sequential gain of a second CSF3R mutation results in G-CSF independence
A new CSF3R mutation, which was acquired at the CSF3R-d715 mutant allele, was found exclusively in the AML blasts and changed a polar threonine residue at amino acid position 595 into a nonpolar isoleucine. This residue is located in a highly conserved threonine-rich region in the extracellular domain of the G-CSF receptor (supplemental Figure 2). Introduction of human CSF3R mutant receptors carrying this new T595I mutation (Figure 3A) into Csf3r-deficient primary mouse BM progenitors resulted in the autonomous outgrowth of myeloid colony-forming cells (Figure 3 and supplemental Table 7). Thus, in the AML phase of disease evolution, 2 different coexisting mutations, the T595I single amino acid substitution and the CSF3R-d715 mutation, had accumulated in the gene encoding the G-CSF receptor. Because expression of the new CSF3R mutant without the δ mutation conferred G-CSF independence, as did the mutant receptor carrying both the δ and the extracellular mutation, this gain of function can be attributed entirely to the T595I mutation. However, the T595I/d715 colonies were bigger than the T595I colonies (supplemental Figure 4), which is suggestive of a higher proliferation capacity by the addition of the CSF3R-d715 mutant.
Functional analysis of CSF3R mutants in myeloid progenitor cell assays. In vitro colony growth of Csf3r-deficient murine hematopoietic progenitor cells expressing different CSF3R mutants. (A) Graphical representation of the different CSF3R constructs. Wild-type (wt), T595I (containing the extracellular mutation at amino acid position 595), d715 (containing the intracellular mutation Q716X, causing the introduction of a stop codon at amino acid position 716), and T595I/d715, containing both mutations as found in the SCN/AML patient. Ig indicates the Ig-like domain; CRH, cytokine receptor homology domain; FNIII, fibronectin type III repeats; TM, transmembrane domain; and cyto, cytoplasmic domain. Nomenclature has been adopted from Layton et al.42 (B) Colonies were grown in the presence of puromycin without growth factor (no GF) or with 100 ng/mL of human G-CSF. The induced colony growth is dependent on the transduction efficiency and the type of CSF3R construct. The transduction efficiency can be deduced from the number of GM-CSF–induced colonies under puromycin selection, because the CSF3R constructs confer puromycin resistance, but do not affect GM-CSF–induced colony growth. Therefore, by dividing the number of colonies by the number of GM-CSF–induced colonies, the transduction efficiency was corrected for.
Functional analysis of CSF3R mutants in myeloid progenitor cell assays. In vitro colony growth of Csf3r-deficient murine hematopoietic progenitor cells expressing different CSF3R mutants. (A) Graphical representation of the different CSF3R constructs. Wild-type (wt), T595I (containing the extracellular mutation at amino acid position 595), d715 (containing the intracellular mutation Q716X, causing the introduction of a stop codon at amino acid position 716), and T595I/d715, containing both mutations as found in the SCN/AML patient. Ig indicates the Ig-like domain; CRH, cytokine receptor homology domain; FNIII, fibronectin type III repeats; TM, transmembrane domain; and cyto, cytoplasmic domain. Nomenclature has been adopted from Layton et al.42 (B) Colonies were grown in the presence of puromycin without growth factor (no GF) or with 100 ng/mL of human G-CSF. The induced colony growth is dependent on the transduction efficiency and the type of CSF3R construct. The transduction efficiency can be deduced from the number of GM-CSF–induced colonies under puromycin selection, because the CSF3R constructs confer puromycin resistance, but do not affect GM-CSF–induced colony growth. Therefore, by dividing the number of colonies by the number of GM-CSF–induced colonies, the transduction efficiency was corrected for.
Discussion
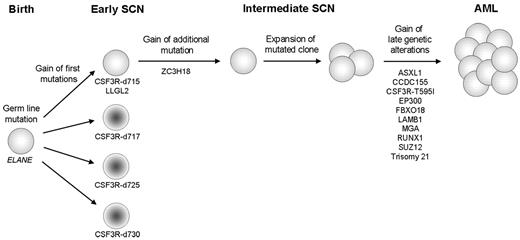
The results of the present study identified nonsynonymous mutations acquired in an SCN patient who progressed to AML. The availability of sequential hematopoietic samples from the childhood SCN phase to overt AML, spanning a period of 17 years, provided the unique opportunity to identify the early and late genetic defects associated with leukemic progression (Figure 4). The CSF3R-d715 mutation and a mutation in LLGL2, which encodes the human homolog of the Drosophila lethal giant larvae (Lgl) gene, were the first 2 acquired mutations in the early SCN phase. Loss of Lgl in Drosophila leads to inadequate distribution of the cell polarity protein Numb, resulting in inappropriate cell-fate determinations and tumor formation in epithelial tissues and the brain.26,–28 In humans, the NUMB protein has been implicated in controlling the balance between symmetric and asymmetric hematopoietic stem-cell divisions. Interestingly, deregulation of NUMB expression has been associated with blast transformation of chronic myeloid leukemia.29,30 How the LLGL2 mutation found in the present study affects hematopoietic stem-cell divisions is still unknown; however, the fact that CSF3R-d715 and LLGL2 mutations were uniformly present in the same myeloid cell could suggest that they cooperate. Hierarchically, the next genetic abnormality occurring in the early SCN phase in the CSF3R-d715– and LLGL2-mutated clone was a mutation in ZC3H18. ZC3H18 is a putative mRNA-binding protein that has an as-yet-unknown function, but has been shown to be essential for differentiation in trypanosomes.31
Schematic representation of the clonal evolution of SCN to AML. The sequential genetic events, starting with the presence of a germline mutation in ELANE are indicated. A sequential gain of CSF3Rδ mutations and an LLGL2 mutation is observed in the early SCN phase. Only the clone harboring the CSF3R-d715 and the LLGL2 mutation gained an additional mutation in ZC3H18, followed by its expansion in the intermediate SCN phase. Gain of 9 additional mutations and trisomy 21 in the mutated population preceded complete transformation to AML. CSF3R-d715-d730 indicates CSF3Rδ mutations at amino acid positions 715-730.
Schematic representation of the clonal evolution of SCN to AML. The sequential genetic events, starting with the presence of a germline mutation in ELANE are indicated. A sequential gain of CSF3Rδ mutations and an LLGL2 mutation is observed in the early SCN phase. Only the clone harboring the CSF3R-d715 and the LLGL2 mutation gained an additional mutation in ZC3H18, followed by its expansion in the intermediate SCN phase. Gain of 9 additional mutations and trisomy 21 in the mutated population preceded complete transformation to AML. CSF3R-d715-d730 indicates CSF3Rδ mutations at amino acid positions 715-730.
In addition, we found small subpopulations harboring distinct CSF3Rδ mutations in the BM at the early SCN stage. All these clones disappeared during the disease course except the CSF3R-d715 clone, which evolved toward AML. The different CSF3Rδ mutations cause expression of distinct truncated G-CSF receptors that all have similar consequences for signaling, resulting in a sustained activation of STAT5.8 STAT5 is a transcription factor implicated in abnormal signaling responses of leukemic cells with mutated forms of the FLT3 receptor (FLT3-ITD) in AML and the BCR-ABL fusion protein in CML.32,33 Furthermore, why one of these CSF3Rδ mutant clones survived in vivo and progressed toward a fully transformed AML clone whereas the other CSF3Rδ variants were extinguished during disease development remains unexplained. However, it is conceivable that the additional mutations in LLGL2 and ZC3H18, which were exclusively present in the CSF3R-d715 clone, conferred a competitive growth advantage of this particular subclone representative of essential early steps in leukemic progression that cooperate with the aberrant signaling from the truncated G-CSF receptor.
In addition to early genetic events, we found 9 mutations that occurred later in the process of leukemic transformation. Of particular interest is the novel CSF3R mutation (T595I), which appeared exclusively in the AML stage and imposed growth factor independence on an already functionally defective G-CSF receptor. A different mutation in the CSF3R transmembrane domain, CSF3R-T617N, which had a similar downstream effect, was found previously to be a constitutive mutation in a family with hereditary chronic neutrophilia and an acquired mutation in 2 AML patients. This mutation is suggested to cause ligand-independent homodimerization and to induce growth factor–independent proliferation and differentiation.34,35 The major difference between the T617N and the T595I mutation in our patient is that the latter was located on the already affected CSF3R-d715 allele, which has been shown to cause increased proliferation and impaired differentiation in cell-line and animal models8,36,37 and which could explain the increase in colony size between the T595I and the T595I/d715 mutants. The acquisition of autonomous growth abilities by myeloid progenitor cells that already express a hyperresponsive G-CSF receptor mutant strongly suggests that perturbed G-CSF signaling was of vital importance for malignant transformation in this case of SCN. To our knowledge, this is the first example of a gain of 2 different mutations in the same receptor in the process of malignant transformation. An important but still open question is whether the administration of G-CSF to this patient contributed to the acquisition of this additional mutation. It is possible that the continuous proliferative pressure imposed by G-CSF on clones carrying mutations in CSF3R-d715 and LLGL2, and later also in ZC3H18, may have provided the context for the selection of a clone harboring this self-activating CSF3R mutation, pushing it to become an autonomously proliferating and dominant leukemic clone.
Abnormalities appearing in the AML phase included mutations in ASXL1, SUZ12, and EP300, all genes encoding proteins involved in chromatin modification. Mutations in ASXL1 have been reported previously in AML and are associated with an unfavorable prognosis.38 SUZ12 is a member of the PRC2 complex that also contains EZH2, the histone methyl transferase responsible for the di- and trimethylation of lysine 27 in the tail of histone 3 (H3K27), imposing a chromatin mark that represses gene expression. Mutations affecting EZH2 and, less frequently SUZ12 have been detected in MDS/MPN patients.25,39,40 In contrast, mutations in EP300 and the highly related CREBBP, encoding histone acetyl transferases that act as transcriptional coactivators, have not yet been reported in myeloid malignancies, but are the most frequent structural abnormalities in follicular lymphoma and diffuse large B-cell lymphoma.23,24 Mutations in CCDC155, encoding coiled-coil domain–containing protein 155 with unknown function; FBXO18, encoding a DNA helicase involved in DNA repair and genomic integrity; LAMB1, encoding an extracellular matrix protein; and MGA, encoding a Max gene associated antagonist of Myc oncoproteins, all represent novel mutations with currently unknown functional significance.
Recurrence is an important criterion used to discriminate driver from passenger mutations in the process of malignant transformation. Interestingly, mutations in CCDC155, LLGL2, MGA, and ZC3H18 were also reported recently by the TCGA consortium in a panel of AML patients (n = 199), albeit at low frequencies. Because frequencies of specific mutations have been shown to vary with the natural history of AML, for example, de novo versus secondary to MDS/MPN or different BM failure syndromes,14,41 it will be of interest to establish how often the newly identified genes are affected in distinct subtypes of secondary AML. Specifically, it will be important to determine whether LLGL2, ZC3H18, or functionally related genes are more generally affected in BM failure syndromes prone to progression to AML and to establish how these mutations contribute to malignant transformation in conjunction with cooperative gene defects.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by the Center of Translational Molecular Medicine, the Dutch Cancer Society KWF kankerbestrijding, and the E-Rare project ELA2-CN.
Authorship
Contribution: R.B. designed and performed the research, collected, analyzed, and interpreted the data, and wrote the manuscript; M.G.V. performed the research and collected the data; M.A.S. contributed analytical tools and analyzed and interpreted the data; P.M.H.v.S. performed the research and collected the data; J.R.H. performed the research; L.B. and W.M.G.-K. collected the samples; A.J.P.V. contributed samples and patient information; P.J.M.V. collected samples and analyzed and interpreted the data; R.G.V. analyzed and provided the TCGA consortium sequencing data; B.L. analyzed and interpreted the data and wrote the manuscript; and I.P.T. supervised the project, collected the samples, designed the research, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ivo P. Touw, PhD, Department of Hematology, Erasmus University Medical Center, Dr Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail: i.touw@erasmusmc.nl.