Oncogenic activating mutations in NOTCH1 occur in more than 50% of T-cell acute lymphoblastic leukemias (T-ALLs). In the present study, we describe a novel mechanism of NOTCH1 activation in T-ALL in which a deletion removing the 5′ portion of NOTCH1 abolishes the negative regulatory control of the extracellular domain and leads to constitutively active NOTCH1 signaling. Polypeptides translated from truncated transcripts encoded by the NOTCH1 deletion allele retain the transmembrane domain of the receptor and are constitutively cleaved by the γ-secretase complex, resulting in high levels of NOTCH1 signaling that can be effectively blocked by γ-secretase inhibitors. Our results expand the spectrum of oncogenic lesions activating NOTCH1 signaling in human T-ALL.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic tumor resulting from the malignant transformation of immature T-cell progenitors.1,2 Activating mutations in NOTCH1 are a hallmark of T-cell transformation and are found in more than half of T-ALL cases.3,4 In the present study, we used array-CGH (aCGH) analysis to investigate the possibility that focal chromosomal rearrangements involving the NOTCH1 locus could also participate in the pathogenesis of human T-ALL. We report an intragenic deletion in NOTCH1 as a novel mechanism of NOTCH1 activation in this disease.
Methods
Patient samples
DNA and lymphoblast samples were provided by the Eastern Cooperative Oncology Group. All samples were collected with informed consent in accordance with the Declaration of Helsinki and with approval from the Columbia University Medical Center institutional review board.
aCGH
aCGH of human T-ALL was performed with SurePrint G3 Human CGH 1 × 1M Oligo Microarrays (Agilent Technologies) using standard procedures.
FISH
FISH was performed by standard methods in interphase nuclei using the RP11-370H5 and CTD-3242B16 BAC probes encompassing the NOTCH1 locus as described previously.5
5′-RACE
5′-Rapid amplification of cDNA ends (5′-RACE) cDNA synthesis was performed with the SMARTer RACE cDNA Amplification Kit (Clontech) using a NOTCH1 exon 28–specific primer (5′-TGGCCTCAGACACTTTGAAGCCCTCAG-3′) and the GC-RICH PCR System (Roche Applied Science).
γ-secretase inhibitor treatments
HEK-293T and HeLa cell cultures were treated for 48 hours with compound E (250nM; Enzo Life Sciences) in DMSO or with vehicle only.
NOTCH1 expression plasmids
The pCDNA3 NOTCH1, pCDNA3 NOTCH1 HD-ΔPEST, pCDNA3 NOTCH1 Jurkat JME17, and pCDNA3 NOTCH1 P12 mutant constructs have been described previously.6,7 The pCDNA3 NOTCH1 N-terminal deletion mutant construct (pCDNA3 NOTCH1 del-N) consists of the ACATGG hexanucleotide, followed by the distal part of the NOTCH1 coding cDNA sequence (RefSeq number NM_017617) starting at position 4998 and a FLAG epitope coding sequence in the 3′ end.
Luciferase reporter assays
NOTCH1 expression plasmids (pCDNA3) were transfected transiently into HeLa cells using FuGene (Roche) together with the pGaLUC RBPJ/CSL luciferase reporter construct. A Renilla luciferase–expressing vector (pRL) was used as an internal control. Luciferase assays were carried out using the Dual Luciferase Assay System (Promega) on a Modulus microplate reader.
Western blotting
Abs against activated NOTCH1 (NOTCH1 Val1774; Cell Signaling Technology), FLAG (DYKDDDDK Tag; Cell Signaling Technology), and actin (SC-1615; Santa Cruz Biotechnology) were used in immunoblot assays using standard procedures.
Results and discussion
A novel 5′ NOTCH1 deletion in human T-ALL
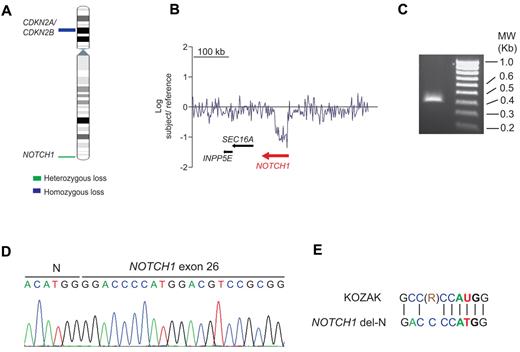
Oncogenic NOTCH1 signaling is a hallmark of T-ALL.3,4,7,,,–11 However, approximately 40% of T-ALL cases lack genetic alterations involving the NOTCH pathway.3,4,7,8 Based on the strict requirement of NOTCH1 for T-cell development12 and the prominent role of NOTCH1 signaling in T-ALL, we hypothesized that as-yet-unrecognized genetic alterations involving NOTCH1 may be implicated in the pathogenesis of T-ALL. To investigate this possibility, we performed aCGH analysis in 69 adult T-ALL samples, which revealed an intragenic deletion encompassing the 5′ region of the NOTCH1 locus in an early cortical double-positive (CD1+, CD4+, CD8+, CD3−) T-ALL patient sample (Figure 1A-B). FISH analysis ruled out the presence of a chromosomal translocation involving NOTCH1 in this sample (data not shown). Detailed analysis of our aCGH data mapped the telomeric breakpoint of this deletion proximal to exon 1 of the NOTCH1 gene and positioned the centromeric deletion breakpoint between exons 24 and 27. Based on these results, we predicted that this focal deletion could result in the expression of an N-terminal truncated form of NOTCH1. To investigate this possibility, we performed 5′-RACE from RNA extracted from NOTCH1-deleted tumor cells using a reverse primer located in exon 28 of NOTCH1. This experiment resulted in the amplification of a 367-nucleotide band consisting of a NOTCH1 del-N allele containing the ACATGG hexanucleotide, followed by the distal part of the NOTCH1 coding cDNA sequence starting in exon 26 at position 4998 (RefSeq number NM_017617; Figure 1C-D). We found that a canonical Kozak consensus sequence flanks a potential NOTCH1 del-N translation initiation site located at nucleotides 14-16, which encode for a methionine corresponding to codon 1668 in the full-length NOTCH1 receptor (Figure 1E). DNA sequencing of exon 34 of NOTCH1 in this patient failed to demonstrate the presence of prototypical PEST domain–truncating mutations, but we did detect 2 nonsynonymous single-nucleotide variants (NOTCH1 C6889A, NOTCH1 T2297P; NOTCH1 G7387T, NOTCH1 A2463T) in the PEST domain region of NOTCH1. Therefore, the predicted NOTCH1 del-N–encoded protein consists of an 887-amino acid polypeptide encompassing the C-terminal HD domain of NOTCH1, followed by the juxtamembrane segment, transmembrane region, and intracellular domains of the receptor.
Isolation and analysis of a NOTCH1 intragenic deletion in human T-ALL. (A) Chromosome 9 ideogram showing deletions identified by aCGH encompassing the CDKN2A/B tumor suppressor gene and the NOTCH1 locus in T-ALL. (B) aCGH plot showing an intragenic focal deletion of NOTCH1 in chromosome band 9q34. (C) Agarose gel electrophoresis of a truncated NOTCH1 cDNA sequence isolated by 5′-RACE using a primer located in exon 28 of NOTCH1. (D) Nucleotide sequence of the NOTCH1 5′ RACE product. (E) Kozak consensus sequence alignment of the NOTCH1 sequences surrounding methionine 1668 located at the N-terminal portion of the NOTCH1 del-N amino acid sequence. R indicates purine (adenosine or guanosine).
Isolation and analysis of a NOTCH1 intragenic deletion in human T-ALL. (A) Chromosome 9 ideogram showing deletions identified by aCGH encompassing the CDKN2A/B tumor suppressor gene and the NOTCH1 locus in T-ALL. (B) aCGH plot showing an intragenic focal deletion of NOTCH1 in chromosome band 9q34. (C) Agarose gel electrophoresis of a truncated NOTCH1 cDNA sequence isolated by 5′-RACE using a primer located in exon 28 of NOTCH1. (D) Nucleotide sequence of the NOTCH1 5′ RACE product. (E) Kozak consensus sequence alignment of the NOTCH1 sequences surrounding methionine 1668 located at the N-terminal portion of the NOTCH1 del-N amino acid sequence. R indicates purine (adenosine or guanosine).
The NOTCH1 del-N mutant is a constitutively active form of NOTCH1 sensitive to γ-secretase inhibitors
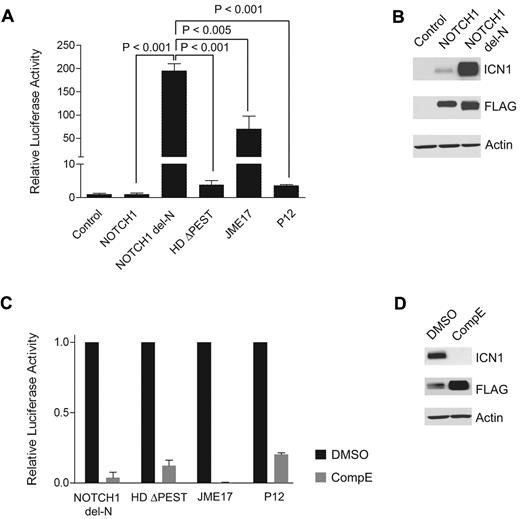
To determine the functional impact of NOTCH1 del-N, we analyzed the levels of NOTCH1 signaling using a RBPJ/CSL luciferase reporter assay in cells transfected with NOTCH1 del-N compared with cells transfected with wild-type NOTCH1 or with a panel of strongly activated NOTCH1 mutants such as NOTCH1 L1601P (HD) ΔPEST,7 NOTCH1 Jurkat JME17,6 and NOTCH1 P12.13 These experiments showed that expression of NOTCH1 del-N results in high levels (200-fold more than wild-type) of activated NOTCH signaling (Figure 2A). NOTCH1 activation by NOTCH1 del-N was significantly higher than that resulting from expression of NOTCH1 HDΔPEST, NOTCH1 Jurkat JME17, or the NOTCH1 P12 mutant allele (Figure 2A). Consistent with the high levels of NOTCH1 activation detected in the luciferase reporter assay, NOTCH1 del-N induced high levels of expression of activated NOTCH1 protein (Figure 2B). We also investigated the ability of compound E, a highly active and specific γ-secretase inhibitor, to inhibit NOTCH1 activation driven by the NOTCH1 del-N allele using a RBPJ/CSL luciferase reporter assay and Western blot analysis. These assays demonstrated complete abrogation of NOTCH1 del-N–driven NOTCH1 signaling and rapid clearance of activated NOTCH1 protein after compound E treatment (Figure 2C-D).
The NOTCH1 del-N mutant shows high levels of NOTCH1 signaling and is sensitive to inhibition of the γ-secretase complex. (A) CSL luciferase reporter assay of ICN1 activity in HeLa cells transfected with empty vector (control) or expressing wild-type NOTCH1 (NOTCH1), NOTCH1 del-N deletion mutant (NOTCH1 del-N), NOTCH1 HD ΔPEST (HDΔPEST), NOTCH1 Jurkat JME17 (JME17), or NOTCH1 P12 (P12) mutant alleles. Reporter activity is shown as the fold change compared with wild-type NOTCH1. Error bars represent SD. P values were derived from Student t test. (B) Western blot analysis of 293T cells expressing empty vector (Control) or FLAG-tagged forms of wild-type and del-N mutant NOTCH1 alleles. ICN1 protein was detected with the NOTCH1 Val1744 Ab, which specifically recognizes the γ-secretase–cleaved activated form of NOTCH1. Total NOTCH1 levels were detected with an anti-FLAG Ab. Actin is shown as a loading control. (C) Transactivation activity of NOTCH1 del-N, HDΔPEST, Jurkat JME17, and P12 NOTCH1 alleles in a CSL luciferase reporter assay in HeLa cells treated with vehicle (DMSO) or a γ-secretase inhibitor (compound E, 250 nM). Reporter activity is shown as the fold reduction compared with vehicle-treated cells for each allele. Error bars represent SD. (D) Western blot analysis of activated NOTCH1 (ICN1) levels in 293T cells treated with compound E (250nM) for 48 hours shows inhibition of NOTCH1 processing and clearance of activated intracellular NOTCH1. ICN1 protein was detected with NOTCH1 Val1744 Ab, total NOTCH1 levels were detected with anti-FLAG Abs, and actin is shown as a loading control.
The NOTCH1 del-N mutant shows high levels of NOTCH1 signaling and is sensitive to inhibition of the γ-secretase complex. (A) CSL luciferase reporter assay of ICN1 activity in HeLa cells transfected with empty vector (control) or expressing wild-type NOTCH1 (NOTCH1), NOTCH1 del-N deletion mutant (NOTCH1 del-N), NOTCH1 HD ΔPEST (HDΔPEST), NOTCH1 Jurkat JME17 (JME17), or NOTCH1 P12 (P12) mutant alleles. Reporter activity is shown as the fold change compared with wild-type NOTCH1. Error bars represent SD. P values were derived from Student t test. (B) Western blot analysis of 293T cells expressing empty vector (Control) or FLAG-tagged forms of wild-type and del-N mutant NOTCH1 alleles. ICN1 protein was detected with the NOTCH1 Val1744 Ab, which specifically recognizes the γ-secretase–cleaved activated form of NOTCH1. Total NOTCH1 levels were detected with an anti-FLAG Ab. Actin is shown as a loading control. (C) Transactivation activity of NOTCH1 del-N, HDΔPEST, Jurkat JME17, and P12 NOTCH1 alleles in a CSL luciferase reporter assay in HeLa cells treated with vehicle (DMSO) or a γ-secretase inhibitor (compound E, 250 nM). Reporter activity is shown as the fold reduction compared with vehicle-treated cells for each allele. Error bars represent SD. (D) Western blot analysis of activated NOTCH1 (ICN1) levels in 293T cells treated with compound E (250nM) for 48 hours shows inhibition of NOTCH1 processing and clearance of activated intracellular NOTCH1. ICN1 protein was detected with NOTCH1 Val1744 Ab, total NOTCH1 levels were detected with anti-FLAG Abs, and actin is shown as a loading control.
Activating mutations in NOTCH1 are present in more than 60% of human T-ALLs, making oncogenic NOTCH1 signaling an attractive target for the development of new, highly specific antileukemic drugs.3,4,7,8 The NOTCH1 del-N T-ALL allele described herein, which results from an intragenic deletion in the NOTCH1 locus and encodes a highly active form of NOTCH1 sensitive to inhibition with γ-secretase inhibitors, expands the repertoire of genetic alterations resulting in aberrant activation of the NOTCH1 signaling pathway in human T-ALL. Similar intragenic deletions in the Notch1 gene resulting in high levels of Notch1 activation and expression of 5′-truncated alternative spliced forms of NOTCH1 have been reported as common oncogenic alterations in mouse models of T-ALL.14,–16 The del-N NOTCH1 allele is reminiscent of the type II Notch1 intragenic deletions described by Ashworth et al.15 Nevertheless, this seems to be a rare event in human T-ALL, because RT-PCR and 5′RACE analysis of T-ALL samples with PEST-only NOTCH1 mutations failed to detect additional samples expressing 5′-truncated forms of NOTCH1 (data not shown).
The identification of NOTCH1 del-N in human T-ALL in the present study highlights the relevance of genomic rearrangements activating NOTCH1 signaling in the pathogenesis of human T-ALL, provides further insight into the mechanisms of NOTCH1-meditated leukemic transformation, and substantiates the value of mouse models of T-ALL harboring activating 5′-truncated forms of Notch1 for the study of T-ALL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Pieter van Vlierberghe for assistance with the aCGH analysis, Dr Vundavalli Murty for FISH analysis, and Dr Tasuku Honjo for the RBPJ/CSL reporter construct.
This study was supported by the Fund for Scientific Research Flanders (to K.D.K.), the Eastern Cooperative Oncology Group tumor bank, and the National Institutes of Health (R01CA120196 to A.F. and U24 CA114737 to E.P.). A.F. is a Leukemia & Lymphoma Society Scholar.
National Institutes of Health
Authorship
Contribution: J.E.H. performed the research and wrote the manuscript; K.D.K. and M.K.D. performed the research; E.P., J.R., P.H.W., and J.M.R. provided patient samples and correlative information; and A.F. designed the study, supervised the research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Adolfo A. Ferrando, Institute for Cancer Genetics, Columbia University, 1130 St Nicholas Ave, Rm 401A, New York, NY 10032; e-mail: af2196@columbia.edu.