Abstract
Transcriptional profiling of differentiating human embryonic stem cells (hESCs) revealed that MIXL1-positive mesodermal precursors were enriched for transcripts encoding the G-protein–coupled APELIN receptor (APLNR). APLNR-positive cells, identified by binding of the fluoresceinated peptide ligand, APELIN (APLN), or an anti-APLNR mAb, were found in both posterior mesoderm and anterior mesendoderm populations and were enriched in hemangioblast colony-forming cells (Bl-CFC). The addition of APLN peptide to the media enhanced the growth of embryoid bodies (EBs), increased the expression of hematoendothelial genes in differentiating hESCs, and increased the frequency of Bl-CFCs by up to 10-fold. Furthermore, APLN peptide also synergized with VEGF to promote the growth of hESC-derived endothelial cells. These studies identified APLN as a novel growth factor for hESC-derived hematopoietic and endothelial cells.
Introduction
During embryogenesis, hematopoietic and vascular lineages arise from extraembryonic and lateral plate mesoderm, derivatives of the posterior and mid regions of the primitive streak (PS), respectively.1 In mouse embryos, the PS appears at embryonic day 6.25 and early stages of mesoderm induction can be tracked by monitoring the expression of key transcription factors such as Brachyury2 and Mixl1,3,4 or cell-surface molecules like Flk-1 and PDGFRα.5,6
Given that the in vitro differentiation of human embryonic stem cells (hESCs) recapitulates aspects of vertebrate development,7 it is not surprising that, analogous to the mouse, genes such as BRACHYURY, MIXL1, KDR (the human Flk-1 homologue), and PDGFRα are expressed during the in vitro mesoderm differentiation of hESCs.8,9 Hemangioblasts, common hematovascular progenitors that emerge from the posterior PS in the mouse embryo,10 also appear in cultures of differentiating mouse11 and human8,9 ESCs after 2-4 days and are followed by the later emergence of primitive erythroid cells, macrophage, and other myeloid and lymphoid lineages.12–15
We have established an in vitro hESC differentiation system to study the development of early hematopoietic mesoderm in a defined, serum-free medium, showing that exogenously added TGFβ-family molecules induce primitive streak-like cells that subsequently differentiate into blood.16–18 To assist in these studies, we modified a hESC line by inserting GFP into the locus of the MIXL1 homeobox gene (MIXL1GFP/w), thus enabling identification and isolation of viable, PS-like cells.8
To discover cell-surface markers and growth factors that would facilitate the identification and generation of hematopoietic lineages, we performed transcriptional profiling during mesoderm induction from MIXL1GFP/w hESCs in cell populations isolated based on their expression of E-CADHERIN (E-CAD), PDGFRα, and GFP. Analysis of these populations demonstrated that GFP+PDGFRα+ and E-CAD−GFP+ cells were highly enriched for the expression of the G-protein–coupled APELIN receptor,19 consistent with the expression of the peptide ligand, APELIN (APLN),20 and its cognate receptor (APLNR) in the PS of the mouse embryo.21 Isolated APLNR+ cell populations were enriched in hemangioblast colony-forming cells (Bl-CFCs), as also shown in a recent report.13 We extended these prior observations by showing that APLNR expression was also observed in MIXL1+ cells representing the anterior primitive streak, a region harboring cells fated to become dorsal mesoderm and endoderm.8,22–24 Furthermore, we showed that APLN enhanced the growth of embryoid bodies (EBs), increased the frequency of hemangioblast colonies, and synergized with VEGF to increase growth of endothelial cells. Collectively, our study demonstrates that APLN is a growth factor that can promote hematopoietic differentiation of hESCs and suggests a hitherto unrecognized role for the APLN/APLNR axis in the regulation of early human hematopoiesis.
Methods
Cell culture and differentiation
The HES3,25 MEL1 (Millipore), H9 (WiCell Research Institute), and MIXL1GFP/w HES38 hESC lines were passaged and differentiated as spin EBs in APEL medium16,17 supplemented with the following growth factor combinations: 100 ng/mL fibroblast growth factor 2 (FGF2); BVS (30 ng/mL bone morphogenetic protein [BMP4], 30 ng/mL VEGF, 40 ng/mL SCF); BloVSAhi (5 ng/mL BMP4, 30 ng/mL VEGF, 40 ng/mL SCF, 100 ng/mL ACTIVIN A); BVhiS (40 ng/mL BMP4, 50 ng/mL VEGF, 25 ng/mL SCF); BVSA (20 ng/mL BMP4, 30 ng/mL VEGF, 40 ng/mL SCF, 20 ng/mL ACTIVIN A); and BVSW (30 ng/mL BMP4, 30 ng/mL VEGF, 40 ng/mL SCF, 100 ng/mL Wnt3a). BMP4, FGF2, VEGF, and SCF were purchased from PeproTech, and ACTIVIN A from R&D Systems. In some cultures, 50 ng/mL APELIN-13 pyroglutamate (APLN; Sigma-Aldrich) was added. Work on hESCs was approved by the Monash University Human Research Ethics Committee.
To identify Bl-CFCs, dissociated day 3 EBs were cultured at 5 to 10 × 103 cells/well in serum-free MethoCult (StemCell Technologies) or in a formulation denoted MC-APEL (1% methylcellulose in APEL medium). MethoCult cultures were supplemented with 30 ng/mL VEGF, 50 ng/mL SCF, and 3 U/mL erythropoietin (EPO) with or without 50 ng/mL APLN, while 10 ng/mL FGF2 was also added to MC-APEL cultures to facilitate the efficient generation of Bl-CFCs.
To bias differentiation toward hematopoiesis, after 4 days in BVS, EBs were cultured in fresh medium with 20 ng/mL BMP4, 50 ng/mL VEGF, 50 ng/mL SCF, 10 ng/mL FGF2, and 30 ng/mL insulin-like growth factor 2 (IGF2).
Endothelial differentiation of hESCs was induced in BVhiS. After sorting day 6 EBs for CD34bright (br)KDRbr progenitors, cells were matured in medium with 50 ng/mL VEGF, with or without APLN addition.
Endodermal differentiation of hESCs was induced in BloVSAhi for 3 days. Cells were sorted based on EPCAM and APLNR expression, reaggregated and cultured further in APEL containing 400 ng/mL noggin (R&D Systems). At day 6, medium was replaced with APEL containing 10−5M retinoic acid (RA). At day 9, the medium was changed to APEL without polyvinyl alcohol (AEL) containing 10−5M RA, 100μM glucagon-like peptide-1, B27, and 10mM nicotinamide. At day 15 of differentiation, EBs were transferred to gelatinized 96-well plates and further maturation was induced in AEL containing 10mM nicotinamide and 50 ng/mL IGF-1.26
Immunofluorescence of adherent cells cultured from hemangioblast colonies
Hemangioblast colonies generated from day 3 EBs were grown for 4 days in methylcellulose, and individual colonies were replated onto gelatinized plates in medium with 30 ng/mL VEGF, 50 ng/mL SCF, and 3 U/mL erythropoietin with or without 50 ng/mL APLN. To assess low-density lipoprotein (LDL) uptake, adherent cells were incubated overnight with 10 μg/mL DiI-labeled acetylated LDL (Molecular Probes), washed in PBS, and imaged. For immunofluorescence, cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, washed in PBS then blocked in PBS/2% FCS/1% goat serum/1% rabbit serum before sequential incubation with mouse anti–human smooth muscle actin (SMA; DAKO), anti–mouse AlexaFluor 488 (Molecular Probes), and 4′,6-diamino-2-phenylindole (DAPI; Sigma-Aldrich). Alternatively, unfixed adherent cells were stained with mouse anti–human CD34 (BD Pharmingen) followed by anti–mouse AlexaFluor 594 and DAPI. Images were captured on a Zeiss Axiovert 200 microscope and processed with Axiovision software (Carl Zeiss).
Flow cytometric analysis
Embryoid bodies were dissociated, and single-cell suspensions were labeled with the following Abs: mouse anti–human E-CADHERIN (Zymed), allophycocyanin-conjugated goat anti–mouse Ig, mouse anti–human PDGFRα, PE-conjugated anti–human IgG, PE-conjugated anti–human EPCAM, PE-conjugated anti–human CD34, allophycocyanin-conjugated anti–human CD33, allophycocyanin-conjugated anti–human CD41a, allophycocyanin-conjugated anti–human GLYCOPHORIN A (CD235a), PE-conjugated anti–human CD45 (all from BD Biosciences), allophycocyanin-conjugated anti–human CD43 (BioLegend), and allophycocyanin-conjugated anti–human APLNR (R&D Systems). Binding and internalization of fluoresceinated APELIN-13 pyroglutamate (APLN-13-Fl; Sigma-Aldrich) was also used to identify APLNR-expressing cells. EBs were cultured with APLN-13-Fl at 400 ng/mL for 4 hours at 37°C before dissociation, followed by labeling with Abs directed to either PDGFRα or EPCAM. Flow cytometric analysis was performed using FACSCalibur, and cell sorting was done using an Influx cell sorter (BD Biosciences).
Microarray analysis
Total RNA was prepared using the High Pure RNA Isolation Kit (Roche Applied Science) according to the manufacturer's instructions. RNA samples were amplified, labeled, and hybridized to Human WG-6 Version 2.0 and HT-12 Version 3 BeadChips (Illumina) by the Australian Genome Research Facility (http://www.agrf.org.au/). Data were analyzed with GenomeStudio Gene Expression Module Version 1.7 (Illumina) using average normalization across all samples. Subsequent data analysis was performed using MultiExperiment Viewer.27 Hierarchical clustering of genes was performed using Pearson correlation with average linkage clustering. Data are available at ArrayExpress (http://www.ebi.ac.uk/microarray_as/ae/) via accession number E-MEXP-3380.
Gene expression analysis
Selected microarray data were validated by quantitative real-time PCR using RNA extracted from the same samples used for microarray analysis. Samples harvested at indicated time points during endoderm differentiation were also analyzed by real-time PCR. TaqMan gene expression probes (Applied Biosystems) are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Relative gene expression levels were calculated as described.18
Morphologic analysis of day 4 differentiating EBs
Day 4 EBs were collected, washed with PBS buffer, and fixed for 20 minutes in 4% paraformaldehyde. Fixed EBs were rinsed in PBS and embedded in 0.7% low-melt agarose and subsequently in paraffin. Histologic sections of day 4 EBs were stained with H&E and images were captured on an Olympus BX51 upright microscope.
Results
The APLNR is highly expressed during mesendodermal differentiation
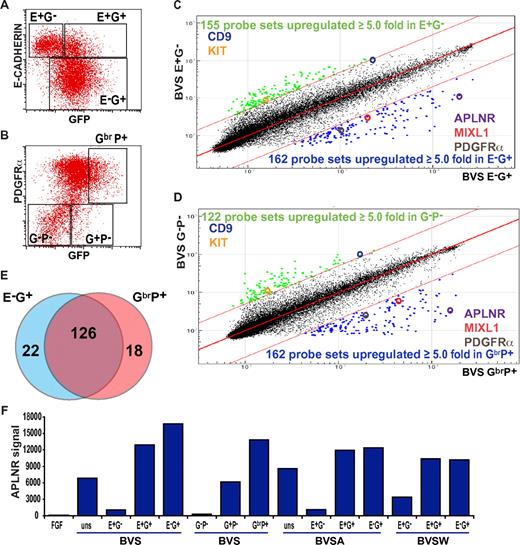
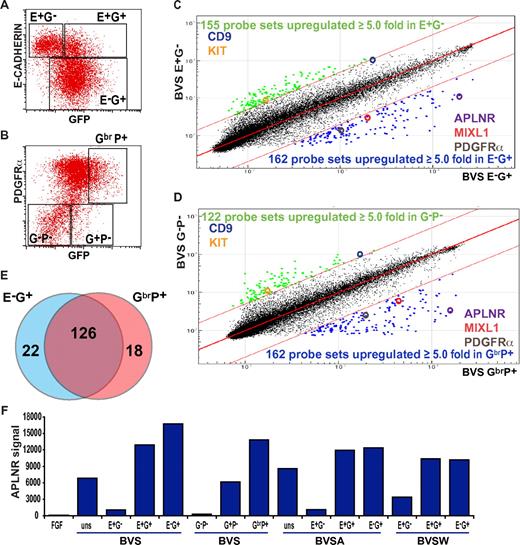
Previous studies in both mouse and human ESCs demonstrated that induction of MIXL1 expression during the early stages of mesoderm differentiation was followed by the reciprocal down-regulation of E-CAHERIN (E-CAD) and up-regulation of PDGFRα expression.8,24 Therefore, to identify additional cell-surface markers of mesodermal precursors, we flow-sorted differentiated GFP-positive, MIXL1-expressing progenitor cells from MIXL1GFP/W hESCs,8 further separating them on the basis of either E-CAD or PDGFRα expression (Figure 1). RNA was extracted from the sorted cell subpopulations, and transcriptional profiles were analyzed using Illumina whole-genome arrays. We identified 162/48 701 probes (148 genes) that were up-regulated at least 5-fold in the E-CAD−GFP+ (E−G+) mesoderm population compared with the least differentiated E-CAD+GFP− (E+G−) cell fraction (Figure 1C). A similar number of probes (162 probes, 144 genes) were differentially expressed in samples derived from an independent experiment in which a GFPbrPDGFRα+ (GbrP+) nascent mesoderm fraction was compared with a less differentiated GFP−PDGFRα− (G−P−) fraction (Figure 1D). Consistent with the reciprocal expression of E-CADHERIN and PDGFRα during mesendodermal induction, 126 (76%) of 166 of the genes up-regulated in the mesoderm fraction were common to both experiments (Figure 1E). In addition to MIXL1 and PDGFRα, the list of genes also included other primitive streak markers such as DKK1, FOXF1, TBX3, EOMES, LHX1, NCAM1, MESP1, MSX1, and members of the HOXB cluster (supplemental Table 2), validating our conclusion that both the GbrP+ and E−G+ fractions marked similar populations.
APLNR expression during mesoderm induction. Flow cytometric analysis of (A) E-CADHERIN and GFP and (B) PDGFRα and GFP expression in MIXL1GFP/w EBs differentiated in medium supplemented with mesodermal-inducing growth factors (BVS). Regions used to sort cells into fractions for further analysis are shown. Comparison of transcriptional profiles of sorted (C) E-CAD+GFP− (E+G−) versus E-CAD−GFP+ (E−G+) cells and (D) GFP−PDGFRα− (G−P−) versus GFPbrPDGFRα+ (GbrP+) cell fractions. Colored dots indicate probes with expression differing by ≥ 5.0-fold from the mean. Several key genes present in each cell population are highlighted. (E) Venn diagram displaying the overlap of genes up-regulated in E−G+ and GbrP+ sorted populations. (F) Relative signal intensities from 4 independent microarray analyses indicating the enrichment of APLNR expression in E−G+, E+G+, G+P+, and GhiP+ nascent mesodermal populations from BVS-, BVSA-, and BVSW-treated EBs (growth factor concentrations provided in “Cell culture and differentiation” and supplemental Figure 1). Samples differentiated under neurectodermal conditions in FGF2 (FGF) served as a negative control for mesoderm differentiation. uns indicates unsorted.
APLNR expression during mesoderm induction. Flow cytometric analysis of (A) E-CADHERIN and GFP and (B) PDGFRα and GFP expression in MIXL1GFP/w EBs differentiated in medium supplemented with mesodermal-inducing growth factors (BVS). Regions used to sort cells into fractions for further analysis are shown. Comparison of transcriptional profiles of sorted (C) E-CAD+GFP− (E+G−) versus E-CAD−GFP+ (E−G+) cells and (D) GFP−PDGFRα− (G−P−) versus GFPbrPDGFRα+ (GbrP+) cell fractions. Colored dots indicate probes with expression differing by ≥ 5.0-fold from the mean. Several key genes present in each cell population are highlighted. (E) Venn diagram displaying the overlap of genes up-regulated in E−G+ and GbrP+ sorted populations. (F) Relative signal intensities from 4 independent microarray analyses indicating the enrichment of APLNR expression in E−G+, E+G+, G+P+, and GhiP+ nascent mesodermal populations from BVS-, BVSA-, and BVSW-treated EBs (growth factor concentrations provided in “Cell culture and differentiation” and supplemental Figure 1). Samples differentiated under neurectodermal conditions in FGF2 (FGF) served as a negative control for mesoderm differentiation. uns indicates unsorted.
In addition to previously characterized mesendodermal genes, we found that the APLNR (also known as AGTRL1/hAPJ),19 a G-protein–coupled receptor for the peptide ligand APELIN (APLN),20,28 was highly enriched in both E−G+ and GbrP+ fractions. A similar selective enrichment for APLNR was observed in 4 independent experiments in which BMP4-based growth factor combinations (supplemental Figure 1) were used to induce GFP+ mesoderm, but APLNR was not expressed in cells differentiated in the presence of FGF2 (a protocol that would bias differentiation toward neuroectoderm29 ; Figure 1F). The increased expression of APLNR in selected cell fractions was confirmed by quantitative (real-time) RT-PCR (supplemental Figure 1).
APLN binds mesodermal and endodermal progenitors in differentiating hESCs
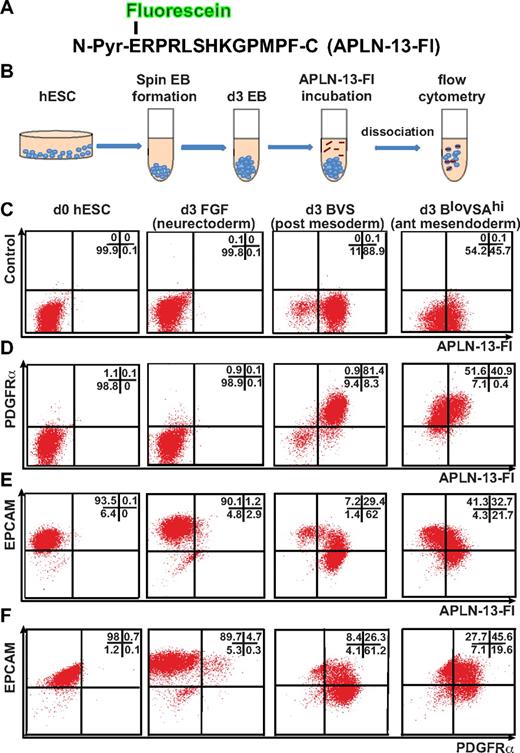
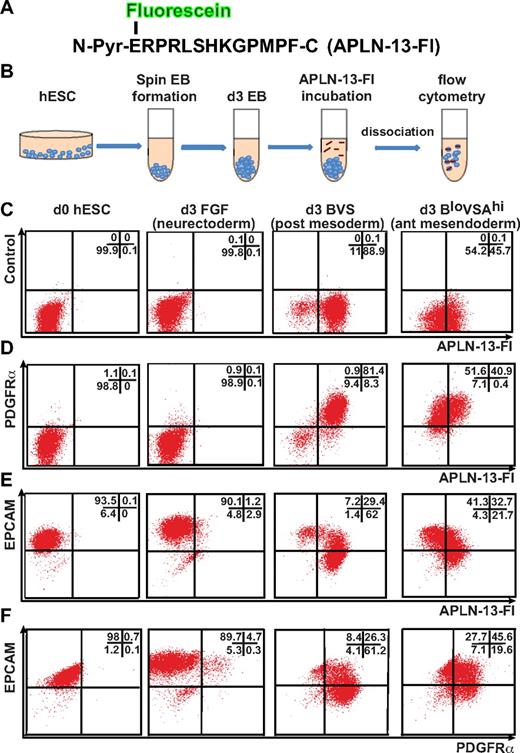
We used 2 independent and complementary methods to evaluate the expression of APLNR on differentiating cells. In the first instance, we identified APLNR-expressing cells by using a fluoresceinated version of its endogenous ligand, APLN. PREPRO-APELIN is a 77-aa protein that is sequentially cleaved to smaller peptides, finally giving rise to an N-terminal pyroglutamated 13-mer, denoted Pyr–APLN-13.20,28 This peptide displays a greater binding efficiency and biologic activity than a longer intermediate, APLN-36.28 Therefore, we synthesized a fluorescein-conjugated APLN-13 N-terminal pyroglutamate peptide (APLN-13–Fl; Figure 2A), based on the previously reported ability of radio-iodinated APLN-13 to bind APLNR+ human endothelial and cardiac cells.30 We empirically determined the optimal duration of APLN-13–Fl incubation before disaggregation of the embryoid bodies to be 4 hours, suggesting that the APLN-13–Fl/APLNR complex was internalized and retained intracellularly over this time. Under these conditions, binding of this peptide proved to be robust method with which to identify APLNR-expressing cell populations during mesodermal induction (Figure 2B). No APLNR expression was detected on undifferentiated hESCs or in cells differentiated in FGF2 for 3 days (Figure 2C).
APLN binds posterior mesodermal and anterior mesendodermal progenitors in differentiating hESCs. (A) Amino acid sequence of synthesized APLN-13 N-terminal pyroglutamate (N-Pyr) peptide conjugated to fluorescein (Fl). (B) Staining protocol used to label cells with APLN-13–Fl. (C-F) Flow cytometric analysis of undifferentiated (d0) hESCs and day 3 EBs differentiated under neurectodermal (FGF), posterior mesodermal (BVS), and anterior mesendodermal (BloVSAhi) conditions labeled with APLN-13–Fl and Abs to PDGFRα or EPCAM. The growth factor concentrations are provided in “Cell culture and differentiation” and supplemental Figure 1. The percentage of cells falling into each quadrant is indicated in top right of each plot.
APLN binds posterior mesodermal and anterior mesendodermal progenitors in differentiating hESCs. (A) Amino acid sequence of synthesized APLN-13 N-terminal pyroglutamate (N-Pyr) peptide conjugated to fluorescein (Fl). (B) Staining protocol used to label cells with APLN-13–Fl. (C-F) Flow cytometric analysis of undifferentiated (d0) hESCs and day 3 EBs differentiated under neurectodermal (FGF), posterior mesodermal (BVS), and anterior mesendodermal (BloVSAhi) conditions labeled with APLN-13–Fl and Abs to PDGFRα or EPCAM. The growth factor concentrations are provided in “Cell culture and differentiation” and supplemental Figure 1. The percentage of cells falling into each quadrant is indicated in top right of each plot.
Flow cytometry demonstrated that day 3 EBs generated using growth factor combinations that biased differentiation toward posterior mesoderm (BVS) generated a greater percentage of APLNR+ cells than growth factor combinations that included a high concentration of ACTIVIN A, which promoted a more anterior mesendodermal fate8,22–24 (BloVSAhi; Figure 2C). Nevertheless, under both conditions, APLNR was coexpressed with PDGFRα, although there was a higher proportion of double-positive cells (> 80%) in BVS-treated EBs (Figure 2D). Conversely, a higher proportion of APLNR+ cells in BloVSAhi treated EBs coexpressed EPCAM, a marker for endodermal precursors as well as for undifferentiated hESCs31 (Figure 2E), suggesting APLNR expression was more strongly up-regulated in posterior mesodermal compared with anterior mesendodermal precursors.
We compared the binding efficiency of the synthesized APLN-13–Fl peptide to a commercially available anti-APLNR Ab by flow cytometry. We found that both detection methods marked similar populations of cells in EBs cultured for 3 days in FGF, BVS, or BloVSAhi (supplemental Figure 2). A greater proportion of the cells were labeled by the APLN-13–Fl than the anti-APLNR Ab (supplemental Figure 2), suggesting this method may be a more sensitive measure of APLNR expression.
Using the anti-APLNR Ab, we compared the expression profiles of MIXL1 and APLNR from days 2 to 6 of differentiation under both posterior mesoderm (BVS) and anterior mesendoderm (BloVSAhi) differentiation conditions. APLNR-expressing cells represented a minority of MIXL1+ cells at day 2, but by day 3 the majority of MIXL1+ cells were also APLNR+ (supplemental Figure 3). This double-positive population was retained until day 6 in BVS-treated cultures but APLNR was rapidly down-regulated after day 4 in high ACTIVIN A–containing cultures. These data relating to the APLNR expression pattern are in accordance with mouse in situ hybridization studies, where APLNR probes labeled the whole primitive streak and adjacent mesoderm but not the endoderm.21 Consistent with our earlier results,8 anterior mesendoderm cells expressed MIXL1 at a higher intensity and for a more prolonged duration (supplemental Figure 3).
Hemangioblast colony-forming cells express APLNR
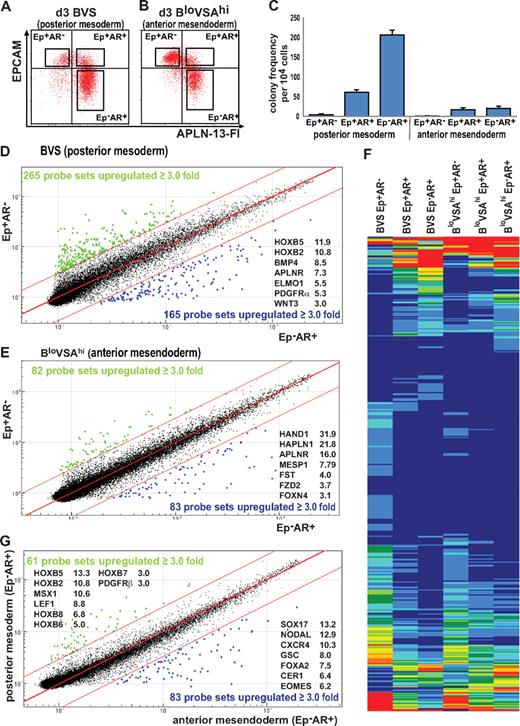
The earliest hematopoietic precursors, Bl-CFC, generate both hematopoietic cells and endothelium.11 They are detected after 2 to 4 days of hESC differentiation,9 and we have previously shown that most Bl-CFCs are contained in a MIXL1+PDGFRα+ fraction of the population.8 Considering the high levels of APLNR expression in day 3 EBs, we examined the frequency of Bl-CFCs in flow-sorted populations from BVS and BloVSAhi-treated cultures (Figure 3A-B; the differentiation potential of these colonies is shown in Figure 6 and supplemental Figure 11). Most colony-forming potential resided in the day 3 BVS-treated EPCAM−APLNR+ (Ep−AR+) cell fraction, with fewer colonies arising from the more immature EPCAM+APLNR+ (Ep+AR+) fraction. In the subpopulations isolated from BloVSAhi cultures, suppression of posterior mesoderm by ACTIVIN A greatly reduced colony frequency (Figure 3C).
Analysis of APLNR- and EPCAM-sorted cells from differentiated EBs. Paired samples of (A) posterior mesodermally (BVS) and (B) anterior mesendodermally (BloVSAhi) differentiated day 3 EBs were flow sorted based on the expression of APLNR and EPCAM expression (boxed areas in the panel). The same gates were used to sort cells differentiated under both protocols to ensure that phenotypically similar populations were compared. (C) Hematopoietic colony-forming potential of sorted cell populations indicating enrichment of hemangioblast colony-forming cells in the APLNR+ fractions (Ep+AR+ and Ep−AR+). Colony frequency was markedly diminished in the anterior mesendoderm differentiations. Error bars represent SEM for n = 3 independent sorting experiments. (D-E) Comparison of microarray-derived gene expression profiles of EPCAM+APLNR− (Ep+AR−) and EPCAM−APLNR+ (Ep−AR+) cells sorted from EBs generated under conditions that favor the formation of (D) posterior mesoderm (BVS) or (E) anterior mesendoderm (BloVSAhi). (F) Heatmap comparing the expression profiles of differentially expressed genes in sorted cell fractions generated under posterior mesodermal (BVS) and anterior mesendodermal (BloVSAhi) conditions, showing similar patterns of expression in both sets of differentiations. (G) Microarray comparison of EPCAM−APLNR+ (Ep−AR+) cell fractions from BVS and BloVSAhi cell fractions representing the most posterior mesoderm and anterior mesendoderm committed fractions. Blue and green dots indicate probes differing by ≥ 3.0-fold from the mean. The fold change of several key genes is indicated for each plot.
Analysis of APLNR- and EPCAM-sorted cells from differentiated EBs. Paired samples of (A) posterior mesodermally (BVS) and (B) anterior mesendodermally (BloVSAhi) differentiated day 3 EBs were flow sorted based on the expression of APLNR and EPCAM expression (boxed areas in the panel). The same gates were used to sort cells differentiated under both protocols to ensure that phenotypically similar populations were compared. (C) Hematopoietic colony-forming potential of sorted cell populations indicating enrichment of hemangioblast colony-forming cells in the APLNR+ fractions (Ep+AR+ and Ep−AR+). Colony frequency was markedly diminished in the anterior mesendoderm differentiations. Error bars represent SEM for n = 3 independent sorting experiments. (D-E) Comparison of microarray-derived gene expression profiles of EPCAM+APLNR− (Ep+AR−) and EPCAM−APLNR+ (Ep−AR+) cells sorted from EBs generated under conditions that favor the formation of (D) posterior mesoderm (BVS) or (E) anterior mesendoderm (BloVSAhi). (F) Heatmap comparing the expression profiles of differentially expressed genes in sorted cell fractions generated under posterior mesodermal (BVS) and anterior mesendodermal (BloVSAhi) conditions, showing similar patterns of expression in both sets of differentiations. (G) Microarray comparison of EPCAM−APLNR+ (Ep−AR+) cell fractions from BVS and BloVSAhi cell fractions representing the most posterior mesoderm and anterior mesendoderm committed fractions. Blue and green dots indicate probes differing by ≥ 3.0-fold from the mean. The fold change of several key genes is indicated for each plot.
Illumina microarray analysis was performed on RNA generated from the sorted posterior mesoderm and anterior mesendoderm fractions (Figure 3). Comparison of the BVS-derived Ep−AR+ fraction with the least differentiated Ep+AR− fraction identified 165 probes (134 genes) that were at least 3-fold up-regulated (Figure 3D). This group included mesodermal markers such as PDGFRα and several HOXB cluster genes (Figure 3D, supplemental Table 3), consistent with the Ep−AR+ fraction marking cells undergoing a transition from primitive streak to nascent mesoderm. A similar comparison between Ep−AR+ and Ep+AR− fractions from BloVSAhi EBs identified 83 probes (66 genes) up-regulated in the Ep−AR+ fraction that contained genes associated with the anterior mesendoderm such as HAND1, FST, FOXN4 (Figure 3E, supplemental Table 4). Comparison of the up-regulated gene lists from both differentiation conditions indicated that 47 of the genes were shared between the two. These shared genes included several primitive streak markers (BMP4, HAND1, IRX3, MSX1 as well as APLNR), consistent with our observation that APLNR marks bipotential mesendoderm. Indeed, the heatmap generated from the 475 probes differentially expressed between Ep−AR+ and Ep+AR− fractions from both posterior mesodermally and anterior mesendodermally differentiated EBs highlighted the overall similarity of gene expression patterns observed under both sets of differentiations as cells progressed from least to most differentiated (Figure 3F, supplemental Table 5).
Therefore, to highlight the differences in outcomes between BVS and BloVSAhi differentiations, we compared the transcriptional profiles of the most differentiated Ep−AR+ samples (Figure 3G). We identified 61 probes (53 genes) that were up-regulated in BVS posterior mesodermally biased APLNR-expressing cells and 83 probes (76 genes) with increased signals in BloVSAhi anterior mesendodermally biased cultures (supplemental Tables 6-7). The genes up-regulated in BVS Ep−AR+ cells included a subset expressed in mesoderm (such as LEF1, MSX1, cardiac ACTIN and HOXB genes) while the BloVSAhi Ep−AR+ cells expressed genes associated with anterior mesendoderm or endoderm lineages (such as SOX17, CER1, FOXA2, GSC). To confirm the ability of APLNR-expressing cells induced in BloVSAhi-differentiated EBs to generate definitive endoderm, we differentiated whole EBs and populations sorted on the basis of EPCAM and APLNR expression toward pancreatic endoderm.26 Endodermally differentiated EBs up-regulated expression of E-CADHERIN and FOXA2, followed by expression of PDX1, AFP, and ALB (supplemental Figure 4). These data suggested that this protocol supported the emergence of both pancreatic and hepatic lineages. Although differentiation of the sorted fractions was less efficient than whole EBs, endodermally differentiated Ep−AR+ cells also expressed both pancreatic and hepatic genes (supplemental Figure 4). These results supported our microarray findings that the expression of APLNR marks both posterior and anterior streak populations.
APLN influences EB size and morphology
To explore the role of APLNR signaling during differentiation, we investigated the effect of APLN addition on hESC differentiation to mesoderm. Spin EBs were differentiated in medium supplemented with BVS, with APLN added on day 2, to coincide with the earliest expression of APLNR (supplemental Figure 3). In the presence of APLN, EBs were consistently larger and developed prominent thin-walled “cysts” that were seldom observed in EBs formed in BVS alone (Figure 4A-C). We confirmed the growth-promoting effect of APLN during mesoderm differentiation with 2 independent hESC lines (Figure 4D, supplemental Figure 5). The increased cell numbers might have resulted from increased cell proliferation and/or a decrease in programmed cell death, given that APLN influences both processes.32,33
APLN alters EB morphology and increases expression of hematopoietic and endothelial genes during mesodermal differentiation. (A-C) Images of H&E-stained paraffin sections showing morphologic appearances of day 4 EBs differentiated in FGF2 alone or in BVS with and without APLN addition at day 2. Several neural rosettes (nr) are evident in the FGF2 EB. EBs that received BVS and APLN were larger with more prominent thin-walled cysts (cy). Scale bar: 100μM. (C) Histogram depicting the mean cell number per EB from day 3 to day 6 under each of these culture conditions. Error bars represent SEM for n = 6 independent experiments. *P < .01 comparing BVS+d2 APLN with BVS at each time point (see also supplemental Figure 5). (E) Histogram indicating the number of genes whose expression was up-regulated or down-regulated (≥ 2.5 fold change) at each time point in EBs cultured with BVS plus a single pulse of APLN at day 2 compared with those receiving BVS alone. (F-I) Histograms comparing the kinetics of gene expression in cells differentiated with or without APLN for selected genes. Panels are grouped to show the profiles of genes whose expression was maximal from day 3 to day 6 as indicated (see also supplemental Figure 6). Abbreviations for gene names are expanded in supplemental Table 8.
APLN alters EB morphology and increases expression of hematopoietic and endothelial genes during mesodermal differentiation. (A-C) Images of H&E-stained paraffin sections showing morphologic appearances of day 4 EBs differentiated in FGF2 alone or in BVS with and without APLN addition at day 2. Several neural rosettes (nr) are evident in the FGF2 EB. EBs that received BVS and APLN were larger with more prominent thin-walled cysts (cy). Scale bar: 100μM. (C) Histogram depicting the mean cell number per EB from day 3 to day 6 under each of these culture conditions. Error bars represent SEM for n = 6 independent experiments. *P < .01 comparing BVS+d2 APLN with BVS at each time point (see also supplemental Figure 5). (E) Histogram indicating the number of genes whose expression was up-regulated or down-regulated (≥ 2.5 fold change) at each time point in EBs cultured with BVS plus a single pulse of APLN at day 2 compared with those receiving BVS alone. (F-I) Histograms comparing the kinetics of gene expression in cells differentiated with or without APLN for selected genes. Panels are grouped to show the profiles of genes whose expression was maximal from day 3 to day 6 as indicated (see also supplemental Figure 6). Abbreviations for gene names are expanded in supplemental Table 8.
APLN increases hematopoietic gene expression
To determine the changes in gene expression accompanying the addition of APLN to the culture medium, we compared the transcriptional profiles of hESCs differentiated in BVS with those of cells that also received a single pulse of APLN at day 2 of differentiation. Differentially transcribed genes (179) were observed from day 3 to day 8 of differentiation and many of the genes up-regulated have been associated with mesoderm development (Figure 4E-I, supplemental Figure 6, supplemental Table 8).
The earliest up-regulated genes included NKD1 (naked cuticle homolog 1), an antagonist of WNT signaling, and the pan-cardiac transcription factor NKX2-5 (Figure 4F). We observed peak expression of transcription factors expressed in hematoendothelial development from day 4 to day 5, including EGR1, GFI1, GATA2, MYC, LYL1, MYB, and SOX18 (Figure 4G-H). Genes involved in angiogenesis that were also up-regulated in response to APLN included CDH5, NOSTRIN, ICAM2, PECAM1, CD34, CD44, and VASH1 (Figure 4, supplemental Figure 6, supplemental Table 8). Up-regulation of the smooth muscle markers TAGLN and TAGLN2 suggested a role for APLN in the generation of other mesodermal components of the vascular wall (Figure 4, supplemental Table 8). At day 5 and day 6, there was a peak in the expression of genes associated with hematopoiesis that included the heme synthesis gene ALAS2, embryonic and fetal globins (HBZ, HBA2, HBE1, HBG1), and TESC, a calcium-binding protein required for megakaryocytic development from human hematopoietic cells.34 These results suggested that major effects of APLN involved the regulation of both blood and endothelial differentiation genes.
APLN augments hematopoiesis
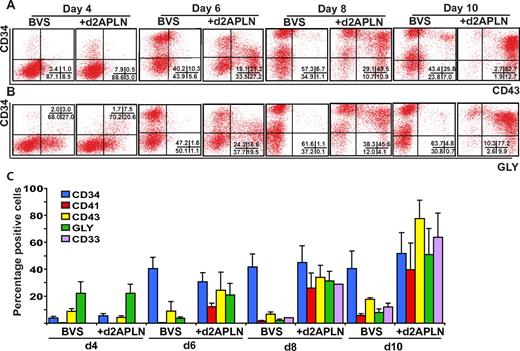
We complemented these gene expression studies by comparing mesodermal and hematopoietic lineage markers on hESC differentiated in BVS with or without APLN from day 2 of differentiation. To bias differentiation toward hematopoiesis, day 4 EBs were cultured in fresh medium with BMP4, VEGF, SCF, FGF2, and IGF2, with or without readdition of APLN. Inclusion of APLN did not affect the expression of mesodermal markers such as MIXL1, PDGFRα, or APLNR for the first 4 days of differentiation (supplemental Figure 7). However, EBs cultured in APLN more rapidly down-regulated PDGFRα expression from day 6 of differentiation. A greater percentage of cells in APLN-supplemented cultures expressed the hematopoietic markers CD41, CD43, CD33, and GLYCOPHORIN A, complementing the rapid reduction in PDGFRα-expressing cells (Figure 5, supplemental Figure 8). Interestingly, the percentage of CD34+ cells did not differ significantly between the treatment groups, indicating that APLN promoted the accumulation of more differentiated hematopoietic cells.
APLN increases expression of hematopoietic markers during hESC differentiation. (A-B) Flow cytometry time-course analysis of CD34, CD43, and GLYCOPHORIN A (GLY) expression on hESCs differentiated in APEL medium supplemented with BVS or BVS with APLN added from day 2. The percentage of cells in each quadrant is shown in the bottom right of each panel. (C) Histogram showing the mean percentage of cells expressing the indicated markers. Error bars represent SEM of 3 independent experiments. Inclusion of APLN resulted in an increased percentage of CD41 (P < .05), CD43 (P < .001), GLYCOPHORIN A (P < .002), and CD33 (P < .05) expressing cells from day 6 of differentiation. Statistical analysis used a 2-way ANOVA with Bonferroni posttest. Detailed FACS plots for all the markers are provided in supplemental Figure 8.
APLN increases expression of hematopoietic markers during hESC differentiation. (A-B) Flow cytometry time-course analysis of CD34, CD43, and GLYCOPHORIN A (GLY) expression on hESCs differentiated in APEL medium supplemented with BVS or BVS with APLN added from day 2. The percentage of cells in each quadrant is shown in the bottom right of each panel. (C) Histogram showing the mean percentage of cells expressing the indicated markers. Error bars represent SEM of 3 independent experiments. Inclusion of APLN resulted in an increased percentage of CD41 (P < .05), CD43 (P < .001), GLYCOPHORIN A (P < .002), and CD33 (P < .05) expressing cells from day 6 of differentiation. Statistical analysis used a 2-way ANOVA with Bonferroni posttest. Detailed FACS plots for all the markers are provided in supplemental Figure 8.
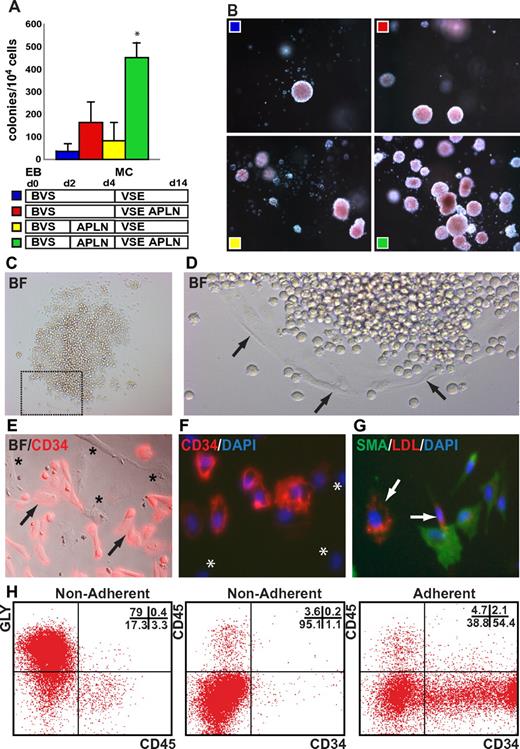
We examined the influence of exogenously added APLN peptide on the frequency of Bl-CFCs. EBs formed from HES3 hESCs differentiated for 3 days in BVS medium were dissociated and seeded into methylcellulose containing VEGF, SCF, and EPO. The addition of APLN to the methylcellulose augmented colony formation at least 3-fold (Figure 6A). Although addition of APLN solely to the initial EB differentiation had little effect on Bl-CFC frequency, the inclusion of APLN from day 2 in both the EB differentiation and the subsequent methylcellulose cultures synergistically augmented the frequency of hemangioblast colony formation, resulting in an ∼ 10-fold increase in colony numbers over non-APLN–supplemented cultures (Figure 6A-B). A similar pattern and magnitude of enhanced colony frequency in response to APLN supplementation was observed with 2 additional independent hESC lines (MEL1 and H9; supplemental Figure 9). Flow cytometric analysis of differentiating hemangioblast colonies harvested from methylcellulose revealed transient expression of CD34 and CD41 and persistent expression of CD43 and GLYCOPHORIN A, consistent with EPO-induced erythroid differentiation. The addition of APLN did not significantly alter the differentiation outcome, suggesting that the major effect of APLN was to increase the frequency of hemangioblast colonies (supplemental Figure 10).
APLN augments hemangioblast colony formation. (A) Augmentation of day 3 hemangioblast colony formation in methylcellulose cultures supplemented with APLN. HES3 hESCs were differentiated as EBs for 3 days in BVS alone or in BVS with APLN added at day 2 (BVS/APLN). Dissociated cells were cultured further in methylcellulose (MC) supplemented with VEGF, SCF, and EPO (VSE) alone or in combination with APLN (VSE/APLN) as indicated. Error bars represent SEM for n = 4 independent experiments; *P < .01 for pairwise comparisons between BVS/APLN VSE/APLN and all other conditions. (B) Dark field images of hematopoietic colonies demonstrating the effect of APLN addition to both the initial mesoderm induction phase (EB) and the methylcellulose (MC) cultures. Original magnification, ×50. (C-D) Bright field images of replated hemangioblast colony demonstrating the presence of both hematopoietic cells and adherent cells (arrows). (E-F) Bright field and immunofluorescence overlay images demonstrating the presence of CD34+ (arrows) as well as CD34− adherent cells (asterisks). (G) Immunofluorescence overlay demonstrating that adherent cells either expressed SMA or took up DiI-Ac LDL (arrows). (H) Flow cytometric analysis of nonadherent and adherent cell fractions from pooled hemangioblast colonies differentiated for 11 days. Samples were stained with Abs to GLYCOPHORIN A (GLY), CD45, and CD34. The percentage of cells falling into each quadrant is indicated in the top right of each plot. Nonadherent cells predominantly comprised GLYCOPHORIN A+ CD45− erythroid cells with a small percentage of CD34−CD45+ myeloid cells whereas over 50% of the adherent cells were CD34+CD45− cells, many of which are likely to be endothelial cells. (G) Approximately 40% of the adherent cells were CD34−CD45− cells, consistent with the proportion of SMA+ smooth muscle cells observed by immunofluorescence (see also supplemental Figure 11). Original magnification: (C) ×50; (D) ×100; (E-G) ×200.
APLN augments hemangioblast colony formation. (A) Augmentation of day 3 hemangioblast colony formation in methylcellulose cultures supplemented with APLN. HES3 hESCs were differentiated as EBs for 3 days in BVS alone or in BVS with APLN added at day 2 (BVS/APLN). Dissociated cells were cultured further in methylcellulose (MC) supplemented with VEGF, SCF, and EPO (VSE) alone or in combination with APLN (VSE/APLN) as indicated. Error bars represent SEM for n = 4 independent experiments; *P < .01 for pairwise comparisons between BVS/APLN VSE/APLN and all other conditions. (B) Dark field images of hematopoietic colonies demonstrating the effect of APLN addition to both the initial mesoderm induction phase (EB) and the methylcellulose (MC) cultures. Original magnification, ×50. (C-D) Bright field images of replated hemangioblast colony demonstrating the presence of both hematopoietic cells and adherent cells (arrows). (E-F) Bright field and immunofluorescence overlay images demonstrating the presence of CD34+ (arrows) as well as CD34− adherent cells (asterisks). (G) Immunofluorescence overlay demonstrating that adherent cells either expressed SMA or took up DiI-Ac LDL (arrows). (H) Flow cytometric analysis of nonadherent and adherent cell fractions from pooled hemangioblast colonies differentiated for 11 days. Samples were stained with Abs to GLYCOPHORIN A (GLY), CD45, and CD34. The percentage of cells falling into each quadrant is indicated in the top right of each plot. Nonadherent cells predominantly comprised GLYCOPHORIN A+ CD45− erythroid cells with a small percentage of CD34−CD45+ myeloid cells whereas over 50% of the adherent cells were CD34+CD45− cells, many of which are likely to be endothelial cells. (G) Approximately 40% of the adherent cells were CD34−CD45− cells, consistent with the proportion of SMA+ smooth muscle cells observed by immunofluorescence (see also supplemental Figure 11). Original magnification: (C) ×50; (D) ×100; (E-G) ×200.
To demonstrate their endothelial differentiation potential, individual hemangioblast colonies from MC-APEL cultures at day 4 of differentiation were transferred to tissue culture–treated plates and cultured for 5-11 days in medium with VEGF, SCF, and EPO, with or without APLN. The majority of colonies (75 of 77; 97%) generated both adherent and floating cells, regardless of the inclusion of APLN (Figure 6C-D, supplemental Figure 11). Immunostaining of these adherent cells indicated the presence of CD34+ endothelial cells, but also a population of CD34− adherent cells (Figure 6E-F, supplemental Figure 11). We examined the ability of the adherent cells to take up LDL, an attribute of endothelial cells, costaining the cultures for expression of the vascular mural marker, SMA. We observed that cells avidly taking up LDL (arrows in Figure 6G and supplemental Figure 11) did not stain strongly for SMA and vice versa, indicating the presence of 2 phenotypically distinct adherent cell populations. Flow cytometric analysis of the differentiated colonies indicated that the nonadherent population was composed of GLYCOPHORIN A+CD45− erythroid cells with very few GLYCOPHORIN A−CD45+ myeloid cells (Figure 6H, supplemental Figure 11), similar to the differentiated cells harvested from methylcellulose (supplemental Figure 10). Conversely, over 50% of the adherent cells were CD34+CD45− cells, consistent with endothelium. However, ∼ 40% of the adherent cells were CD34−CD45− cells, consistent with the proportion of SMA+ smooth muscle cells observed by immunofluorescence (Figure 6H, supplemental Figure 11). The demonstration that most hemangioblast colonies generated hematopoietic cells, endothelium, and smooth muscle is reminiscent of a similar tripotential Bl-CFC isolated from the early mouse embryo.10 As far as we are aware, tripotential differentiation has not previously been documented for human Bl-CFCs.
When colony-forming assays were performed using human CD34+ cord blood cells, the addition of APLN did not alter colony frequency or morphology, consistent with our finding that APLNR was not expressed on CD34+ cord blood cells (supplemental Figure 12).
APLN enhances the endothelial growth and hematopoietic colony-forming potential of day 6 CD34brKDRbr cells
Both APLNR and APLN are highly expressed in endothelial precursors of the nascent vasculature and play a role during angiogenesis in regulating blood vessel diameter.35–38 Therefore, we investigated the expression of APLNR and APLN during the differentiation of endothelial progenitors from hESCs.
Cells coexpressing high levels of CD34 and KDR are enriched for endothelial progenitors in adults39 and endothelial progenitors from differentiating hESCs also express high levels of CD3440 and KDR (M.C., E.G.S., and A.G.E., unpublished results, March 2010). The APLNR was expressed in CD34brKDRbr cells sorted from day 6 EBs differentiated in medium with BVhiS (Figure 7A). APLNR expression was maintained when these cells were cultured for a further 6 days in VEGF, but was absent from a more mature source of endothelium, HUVECs, consistent with previous reports37 (Figure 7B). Although day 6 CD34brKDRbr cells expressed high levels of the TIE2/TEK receptor and its ligand, ANGPT1, which has been shown to induce APLN expression in HUVECS,37 transcripts for APLN were initially absent in CD34brKDRbr cells (Figure 7C-D). However, during the 6-day period in culture, APLN transcription was markedly up-regulated, associated with high levels of expression of ANGPT2 and a down-regulation of the TIE2/TEK receptor (Figure 7E-F). Transcripts for CD34 and KDR were retained by the hESC-derived endothelial cells in contrast to HUVECs, which only expressed low levels of these genes (Figure 7G-H).
APLN enhances the growth of endothelial and hematopoietic cells from CD34brKDRbr progenitors. (A) Flow cytometric analysis of KDR and CD34 expression in day 6 EBs differentiated under endothelium-inducing growth conditions (BVhiS). The CD34brKDRbr sorted cell population is indicated by the boxed area. (B-H) Microarray expression data for the indicated genes in undifferentiated hESC cells (hESC), day 6 CD34brKDRbr sorted progenitor cells (d6), CD34brKDRbr cells cultured in vitro for an additional 6 days (d6 + 6), and HUVEC cells. (I) Histogram showing the fold expansion after culturing 1.5 × 104 sorted day 6 CD34brKDRbr cells for 8 days in medium supplemented with the indicated concentrations (ng/mL) of VEGF and APLN. Error bars represent SEM for n = 5 independent experiments. *P < .0005 for VEGF/APLN vs VEGF. (J) Histogram showing the mean frequency of hematopoietic colony-forming cells in methylcellulose cultures seeded with day 6 sorted CD34brKDRbr progenitor cells, cultured with VEGF, SCF, and EPO in the presence or absence of 50 ng/mL APLN. Error bars represent SEM for n = 6 independent experiments. *P < .03 for APLN addition.
APLN enhances the growth of endothelial and hematopoietic cells from CD34brKDRbr progenitors. (A) Flow cytometric analysis of KDR and CD34 expression in day 6 EBs differentiated under endothelium-inducing growth conditions (BVhiS). The CD34brKDRbr sorted cell population is indicated by the boxed area. (B-H) Microarray expression data for the indicated genes in undifferentiated hESC cells (hESC), day 6 CD34brKDRbr sorted progenitor cells (d6), CD34brKDRbr cells cultured in vitro for an additional 6 days (d6 + 6), and HUVEC cells. (I) Histogram showing the fold expansion after culturing 1.5 × 104 sorted day 6 CD34brKDRbr cells for 8 days in medium supplemented with the indicated concentrations (ng/mL) of VEGF and APLN. Error bars represent SEM for n = 5 independent experiments. *P < .0005 for VEGF/APLN vs VEGF. (J) Histogram showing the mean frequency of hematopoietic colony-forming cells in methylcellulose cultures seeded with day 6 sorted CD34brKDRbr progenitor cells, cultured with VEGF, SCF, and EPO in the presence or absence of 50 ng/mL APLN. Error bars represent SEM for n = 6 independent experiments. *P < .03 for APLN addition.
We evaluated the ability of APLN to augment endothelial growth. While APLN alone did not promote expansion of sorted day 6 CD34brKDRbr cells, the inclusion of APLN significantly enhanced the endothelial growth-promoting effects of VEGF (Figure 7I), a result reminiscent of the observation that the combination of Apln and Vegf induced larger vessels in a mouse model of angiogenesis.36 Finally, we also demonstrated that the same day 6 CD34brKDRbr population included committed hematopoietic CFCs that were responsive to APLN (Figure 7J).
Discussion
We have defined a novel role for APLN, the peptide ligand for the APLNR, during the in vitro hematopoietic differentiation of hESCs. The combination of increased EB size and enhanced frequency of Bl-CFC in response to APLN resulted in an ∼ 10-fold increase in the yield of colony-forming cells, documented in 3 independent hESC lines. The inclusion of APLN from day 2 of differentiation enhanced the generation of hematopoietic cells from differentiating hESC and hematopoietic CFCs present in sorted day 6 CD34brKDRbr cells retained APLN responsiveness. We also showed that treatment of the APLNR-expressing CD34brKDRbr progenitors with APLN and VEGF synergistically promoted endothelial cell growth. This role for APLN in human hematopoiesis complements its cardiovascular actions in adult tissues, in which APLN antagonizes the pressor functions of vasopressin, stimulates water intake, induces arterial vasodilation in a nitric oxide–dependent fashion, and has positive inotropic effects on the heart.41–43
The earliest observable effects of APLN coincided with a period of differentiation during which its sole receptor, APLNR, was up-regulated on MIXL1+ mesendoderm, consistent with reports documenting APLNR expression during gastrulation in the frog, zebrafish, and mouse.21,44–47 Vodyanik and colleagues also recently identified APLNR expression on hESCs differentiated by coculture with mouse OP9 cells.13 In their study, the absence of FOXA2 expression in APLNR+ cells coupled with the ability of this population to generate mesenchymal and hemangioblast colonies suggested APLNR might be restricted to progenitors of lateral plate and extraembryonic mesoderm.13 While we also identified APLNR expression on mesodermal populations, our data showed that hESCs differentiated toward more anterior primitive streak derivatives, including endoderm, also transiently expressed the APLNR. This conclusion was supported by the observation that endoderm-associated genes such as SOX17, NODAL, GSC, and FOXA2 were up-regulated in APLNR+ cells generated in medium containing high levels of ACTIVIN A. Under such conditions, we observed a concomitant decrease in the Bl-CFC frequency of the APLNR+ population, consistent with ACTIVIN A–mediated suppression of posterior mesoderm. Finally, we showed that sorted APLNR+ populations could be further differentiated to cells expressing pancreatic and hepatic endoderm genes. Taken together, our results suggest that early APLNR expression marks a broad spectrum of primitive streak-like cells during hESC differentiation, consistent with its expression throughout the entire length of the primitive streak of the gastrulating mouse embryo.21
In all species examined thus far, the APLNR homologue is expressed in mesendodermal precursors from which cardiovascular and hematopoietic lineages subsequently develop.21,44–47 Later in embryogenesis, mouse Aplnr is highly expressed on endothelial cells undergoing angiogenesis, especially at the tips of intersomitic vessels, and on endothelial cells in the AGM region, on vascular smooth muscle, and on the myocardium.37,38,42 Similar patterns of cardiovascular expression are seen in the zebrafish and frog.35,47,48 Analysis of transcriptional profiles of mesodermally biased differentiating hESC cultures pulsed with APLN at day 2 of differentiation revealed a subsequent up-regulation in transcription of genes associated with hematoendothelial differentiation. Given the documented expression of APLNR in the midgestation myocardium,42 it was noteworthy that APLN-treated cultures also transiently up-regulated the WNT antagonist NKD1 and the pan-cardiac transcription factor NKX2.5.
In the context of these findings, it is relevant that morpholino knockdown of Xapelin or Xmsr (the Xenopus APLNR homologue) in the frog embryo impaired blood, endothelial, and cardiac development.44 Conversely, embryos injected with Xpreproapelin displayed expanded domains of endothelial and hematopoietic gene expression.44 Cox et al similarly observed that a localized source of Apln stimulated endothelial outgrowth and that Xapelin and Xmsr morpholinos led to reduced and disordered endothelial development.35
In the zebrafish, embryos mutant for either of 2 redundantly acting APLNR-like genes (Agtrla and Agtrlb) displayed abnormal cardiac development as the dominant phenotype.45,47 As in Xenopus, overexpression of Apln also perturbed myocardial development in zebrafish probably by affecting cell migration, but hematopoietic and vascular abnormalities were not described.44,45,47 However, the Apln knockout mouse exhibited a mild phenotype characterized by a reduced diameter of intersomitic vessels during embryogenesis, narrow blood vessels in the trachea and skin postnatally, and impaired recovery from hind-limb ischemia, consistent with the role for Apln in angiogenesis.36,37,49 In one study examining the phenotype of Aplnr deleted mice, null mutants were born at close to the expected Mendelian frequency (19%) but displayed an increased vasopressor response to angiotensin II.50 Conversely, 2 other groups reported a more severe phenotype with reduced numbers of viable homozygous Aplnr−/− embryos.43,49 Although details of the embryonic mutants have not been published, surviving Apln−/− and Aplnr−/− mice displayed decreased exercise capacity and impaired sarcomeric function in isolated cardiomyocytes.49 In another study, Aplnr−/− mice displayed reduced water intake and were unable to concentrate urine in response to water deprivation.43 To date, no hematopoietic phenotype has been reported in surviving Apln−/− or Aplnr−/− mice. As such, it is possible that other signaling systems operating during the critical period of hematopoietic progenitor specification are able to compensate for loss of both APLN and its receptor. In this context, given that APLN synergized with VEGF in promoting endothelial cell growth from hESCs, it would be of interest to examine the effect of APLNR signaling loss under conditions in which VEGF signaling was compromised.
In conclusion, our observations indicate a previously unrecognized role for APLNR signaling in regulating hematopoiesis in differentiating hESCs. Early hematopoietic progenitors express APLNR and the inclusion of APLN peptide in culture media enhances the efficiency of blood cell formation from differentiating hESCs. It is tempting to speculate that APLN might play a role in the generation of hematopoietic cells from endothelium, given the documented expression of APLNR in AGM vasculature on the mouse embryo.38 The molecular cascades downstream of APLN/APLNR signaling that mediate these hematopoietic effects remain to be elucidated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Robyn Mayberry and the staff of StemCore Vic for provision of hESCs, David Haylock and Susie Nilsson for provision of CD34+ cells purified from human umbilical cord blood, and Andrew Fryga and the staff of FlowCore for cell sorting.
This work was supported by grants from the Australian Stem Cell Center, Stem Cells Australia, the Juvenile Diabetes Research Foundation, and the National Health and Medical Research Council of Australia (NHMRC).
A.G.E. and E.G.S. are Senior Research Fellows of the NHMRC.
Authorship
Contribution: Q.C.Y., C.E.H., M.C., E.S.N., J.V.S., and K.G. performed the research and analyzed data; and Q.C.Y., C.E.H., E.G.S., and A.G.E. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew G. Elefanty, Monash Immunology and Stem Cell Laboratories, Building 75, STRIP 1, West Ring Road, Monash University, Clayton, Victoria 3800, Australia; e-mail: andrew.elefanty@monash.edu.
References
Author notes
Q.C.Y. and C.E.H. contributed equally to this study.