Abstract
Herein we describe that in classic Hodgkin lymphomas (cHL, n = 25) the lymph node (LN) stroma displayed in situ high levels of transcription and expression of the disulfide-isomerase ERp5 and of the disintegrin-metalloproteinase ADAM10, able to shed the ligands for NKG2D (NKG2D-L) from the cell membrane. These enzymes were detected both in LN mesenchymal stromal cells (MSCs) and in Reed-Sternberg (RS) cells; in addition, MIC-A and ULBP3 were present in culture supernatants of LN MSCs or RS cells. NKG2D-L–negative RS cells could not be killed by CD8+αβT or γδT cells; tumor cell killing was partially restored by treating RS cells with valproic acid, which enhanced NKG2D-L surface expression. Upon coculture with LN MSCs, CD8+αβT and γδT cells strongly reduced their cytolytic activity against NKG2D-L+ targets; this seems to be the result of TGF-β, present at the tumor site, produced in vitro by LN MSCs and able to down-regulate the expression of NKG2D on T lymphocytes. In addition, CD8+αβT and γδT cells from the lymph nodes of cHL patients, cocultured in vitro with LN MSCs, underwent TGF-β–mediated down regulation of NKG2D. Thus, in cHL the tumor microenvironment is prone to inhibit the development of an efficient antitumor response.
Introduction
It is now accepted that the so-called stress surveillance contributes to the anti–neoplastic immunity, both in solid tumors and hematologic malignancies, through the activation of the NKG2D receptor that recognizes NKG2D ligands (NKG2D-L) on cancer cells, including the MHC class-I related chain-A and -B (MIC-A/B) and the UL16-binding proteins 1-4 (ULBPs).1-5 These ligands are commonly expressed at very low levels or retained in the cytoplasm, in healthy tissues, but their transcription and surface expression are enhanced on viral infection or tumor transformation.5-9 Besides natural killer cells and CD8+ T lymphocytes, γδ T cells can recognize these molecules and activate an antitumor response in different cancers.10-17 In this regard, we have described that γδ T lymphocytes belonging to the Vδ1 subset are expanded in patients with chronic lymphocytic leukemia (CLL) and non-Hodgkin lymphomas (NHLs), where they proliferate in response to tumor cells, provided they express NKG2D-L, exert cytotoxicity, and produce anti-neoplastic or pro-differentiating cytokines, such as TNF-α and IL-4.15,16 However, NKG2D-L can be shed by tumor cells and, in their soluble form, interact with NKG2D expressed by effector lymphocytes and hinder the recognition of tumor cells.18,19 Proteolytic cleavage of MIC-A has been shown to depend on the thiol isomerase ERp5 and the disintegrins and metalloproteinases ADAM10 and ADAM17, which are also able to cleave ULBPs.19,22 Overexpression of these enzymes has been reported in multiple myeloma and other tumors.19-23 In turn, soluble (s) NKG2D-L and cytokines produced at the tumor site can down-regulate the expression of the NKG2D receptor on effector lymphocytes, contributing to tumor escape from immunosurveillance.22-25 Indeed, the TGF-β has been shown to reduce the surface density of the NKG2D receptor on CD8+ T and NK cells, impairing their antitumor reactivity in cancer patients.24-26 Moreover, we and others reported that plasma levels of sNKG2D-L correlate with disease progression in multiple myeloma, CLL, NHL, and acute myeloid leukemias; in particular, among sNKG2D-L, both sMIC-A and sULBP2 have been shown as a prognostic marker for multiple myeloma and for the identification of early-stage CLL patients with risk of disease progression.14-16,21,27,28
In this paper, we studied 25 classic HL (cHL) and found that the tumor microenvironment is prone to inhibit the development of an antitumor response. This is mainly because of the release of soluble MIC-A and ULBP3 by lymph node mesenchymal stromal cells (LN MSCs) and Reed-Sternberg (RS) cells, which display high expression of ERp5 and ADAM10, and to the production of TGF-β by LN MSCs, leading to NKG2D down-regulation on effector lymphocytes.
Methods
Patients
Twenty-five patients were analyzed between January 2009 and December 2010, diagnosed with classic Hodgkin lymphoma (cHL, 15 nodular sclerosis [NS], and 10 mixed cellularity [MC]), according to the WHO classification, at the Department of Oncology and Hematology, University of Genoa.29 LN bioptic specimens were obtained under conventional diagnostic procedures, provided informed consent in accordance with the Declaration of Helsinki, and the study was approved by the University of Genoa institutional ethical committee (institutional review board approval 0026910/07, renewal March 2009). Their clinical, histologic, and molecular features are summarized in Table 1. Paraffin-embedded LN samples used for diagnosis and for histochemistry and molecular studies were processed at the Pathology Unit of the IRCCS AOU San Martino–IST National for Cancer Research in Genoa. Fifteen healthy LNs, taken as sentinel LN during surgical approaches and resulted free of neoplastic disease, were also studied.
mAbs and reagents
The FITC-conjugated or the PE-conjugated or allophycocyanin-conjugated (APC) anti–CD8 mAb, the APC or FITC-anti–CD3 mAb, the anti-CD15, the FITC-anti–IL-10 mAbs were from BD Biosciences PharMingen Europe. The anti–CD30 mAb was from Ventana Medical System, and the anti–HLA class-I W6/32 (IgG2a), the anti-SH2 (CD105, IgG1), the anti-SH3 (CD73a, IgG2b), the anti-SH4 (CD73b, IgG1), the anti-CD34 (IgG1), producing hybridomas were purchased from ATCC. The anti–prolyl-4-hydroxylase mAb (clone 5B5, IgG1) was purchased from Dako Italy. The anti-ADAM10 mAb (MAB1427), the anti-NKG2D (MAB139) mAb, and the Ig-NKG2D chimera (NKG2D-Fc) were purchased from R&D Systems, the anti–trans-glutaminase (TG2) from NeoMarkers, the anti–IL-10, the anti–TGF-β, the rabbit polyclonal anti-PDIA6 (ERp5), the rabbit polyclonal anti-ADAM10, and the FITC-goat anti–rabbit (GAR) antisera were from Abcam, and the anti–Vδ1 mAb A13 and the anti–Vδ2 mAb BB3 (both IgG1) were prepared as described.15 The anti–MIC-A mAbs AMO1 and BAMO3 were from Immatics Biotechnologies, and the anti–ULBPs mAbs (anti–huULBP1 M291, anti–huULBP2 M311, anti–huULBP3 M551, and anti–huULBP4 M475) were kindly provided by Amgen (MTA no. 200309766-001). Complete medium was composed of RPMI 1640 (Biochrom) with 10% FCS (Biochrom) supplemented with penicillin, streptomycin, and l-glutamine (Biochrom). The anti–vimentin and anti–collagen mAbs, all-trans-retinoic acid (ATRA) and valproic acid (VPA) were from Sigma-Aldrich, recombinant IL-2 and IL-15 (rIL-2, rIL-15) were from PeproTech.
ELISA for MIC-A, ULBPs, and TGF-β
Soluble (s)MIC-A, sULBP2, and sULBP3 were measured in supernatants (SNs) by ELISA as described. The anti–MIC-A mAbs AMO1 and BAMO3 were from Immatics Biotechnologies, and the anti-ULBPs mAbs (anti–ULBP2 anti–ULBP2 M311, IgG1; anti–ULBP3 M551, IgG1) were provided by Amgen. The anti-ULBP2 and anti-ULBP3 detection mAbs (MAB1298, IgG2a; MAB15171, IgG2a) were from R&D Systems. Anti–mouse IgG2a HRP was from Southern Biotechnology Associates. Plates were developed with 2,2′azinobis(2ethylbenzothiazoline-6-sulfonic acid; Sigma-Aldrich) and read at a OD450 nm. Results are expressed as nanograms per milliliter and referred to a standard curve obtained with the MIC-A/Fc, ULBP2/Fc, or ULBP3/Fc chimeras (R&D Systems).15 TGF-β1 was measured after treatment for 1 hour of each SN with 1N HCl followed by 1N NaOH with a TGF-β1 specific kit (Bender MedSystem). Results were normalized to a standard curve and expressed as pg/mL.
Isolation and culture of LN MSCs and coculture with T cells
LN MSCs were obtained by culturing LN cell suspensions from cHL patients in 6-well plates (5 × 106 cells/well) in MEM-α (GIBCO) complete medium.30 After 3 days, nonadherent cells were washed away and adherent cells cultured for additional 7 days. LN MSCs expressed HLA-I, SH3/CD73a, CD90, SH2/CD105, prolyl-4-hydroxylase (PH4), collagen, vimentin, transglutaminase (TG; “ERp5+/ADAM10 LN MSCs and RS cells can be isolated from cHL: shedding of MIC-A and ULBP3”), bone sialoprotein, osteopontin, SH4/CD73b, CD44, β1-integrin/CD29, ICAM1/CD54, alkaline phoshatase, but not CD45, CD31, CD34, CD33, CD3, CD2, CD16, CD14, ICAM2, ICAM3, CD80, CD86, CD83, and HLA-DR (not shown).
CD8+αβT- and γδT-cell populations were obtained from heparinized blood of healthy donors.31 CD8+αβT cells were separated from whole blood samples with the RosetteSep isolation kit for CD8+ T cells (StemCell Technologies) according to the manufacturer's instructions. By applying this procedure, the starting CD8+αβT cells were more than 98% pure (n = 15 experiments). To obtain γδ T cells, peripheral blood mononuclear cells were isolated by Ficoll Hypaque density gradient centrifugation of blood samples from the same donors. γδ T cells were purified with the Minimacs separation kit (Miltenyi Biotec) specific for γδT cells according to the manufacturer's instructions. After this separation, γδT cells were always more than 96%. Vδ1 and Vδ2 peripheral T cells were separated from peripheral blood mononuclear cells using home-made anti-Vδ1 (A13) or anti-Vδ2 (BB3) mAbs and EasySep custom kit (Stem Cell Technologies) according to the manufacturer's instructions. The purity of Vδ1 or Vδ2 T cell population was always more than 95%. All the cell populations were stimulated with 0.5 μg/mL of PHA in the presence of 10 ng/mL of IL-2 in 96U-bottomed microwells; after 15 days of culture, CD8+αβT cells were always 99% to 100% (n = 15), and more than 98% of cells were γδ T cells (n = 15). CD8+αβ or γδ T cells were immediately used in coculture experiments with LN MSCs. These cocultures were performed using a different ratio between LN MSCs and responder lymphocytes (1:10 to 1:80) and evaluated at different time points (from 12 hours to 8 days). In some experiments, rIL-15 was added (10 ng/mL), in the absence or presence of the anti–TGF-β mAbs (5 μg/mL).
Immunofluorescence and cytofluorimetric analysis
Immunofluorescence on cells isolated from lymph nodes or on cultured cells was performed with the various mAbs indicated above labeled with the fluorochrome AlexaFluor-488 (from now on indicated as FITC) or PE, or AlexaFluor-647 contained in the Zenon Tricolor Labeling Kit for mouse IgG1 (Invitrogen), followed by mAbs to the indicated markers labeled with the appropriate fluorochromes. Control aliquots were stained with AlexaFluor-labeled isotype-matched irrelevant mAbs. For intracytoplasmic staining, cells were washed with PBS, NaN3 0.1% and FCS 0.5%, fixed (3% paraformaldehyde in PBS, 5 minutes at 4°C), and permeabilized (1% Nonidet-P40 in PBS, 5 minutes at 4°C). After washing, samples were stained with the indicated mAbs or polyclonal antibodies, for 30 minutes at 4°C, followed by anti–isotype goat anti–mouse (GAM) or GAR antiserum conjugated with the appropriate fluorochromes and analyzed by CyAn ADP flow cytometer (Beckman Coulter). Results are expressed as log of mean fluorescence intensity (MFI) or percentage of positive cells, or as the ratio between the MFI of each sample and the negative control, as indicated in the figure legend.
Cytotoxicity assay
Cytolytic activity of CD8+αβT and γδT cells was analyzed in a 4-hour 51Cr-release assay against the RS773 cell line obtained from the LN of a cHL, or the L-540, KM-H2, L-428, and HDLM-2 cell lines obtained from pleural effusions of cHL patients and purchased from DSMZ GmbH (Braunschweig), labeled with 51Cr, at an E:T ratio of 10:1, in V-bottomed microwells. A total of 100 μL of supernatant was measured in a γ-counter, and the percentage of 51Cr-specific release was calculated as described previously. Some experiments were done using as targets the RS773, the L-540, KM-H2, L-428, and HDLM-2 cell lines pretreated for 3 days with 2.5mM VPA or with 10μM ATRA; in some samples, the effector cells were exposed to saturating amounts (3 μg/mL) of the anti-NKG2D, the anti–MIC-A, the anti-ULBP3 mAb, or the Ig-NKG2D chimera before the cytotoxicity assay.15 In other experiments, CD8+αβ or γδ T cells were used as effector cells after coculture with LN MSCs.
Immunohistochemistry
Paraffin-embedded LN samples from 18 cHL patients (9 NS and 9 MC) and 6 normal LNs, obtained under diagnostic procedures, were analyzed for the expression in situ of MIC-A, ULBPs, Erp5, ADAM10, and TG. Immunohistochemistry was performed on 4-μm-thin sections, deparaffinized, and treated with methanol 3% hydrogen peroxide in methanol for 10 minutes to quench endogenous peroxidase. Antigen retrieval was performed with sample incubation in Triton-X 100 0.1% for 10 minutes at room temperature for the rabbit polyclonal anti-PDIA6 (ERp5) or with boiling citrate buffer 10mM, pH 6, for the rabbit polyclonal anti-ADAM10, the anti-TG2 mAb, the anti-CD30, anti–MIC-A, and anti-ULBP3 mAbs. The various antibodies were added at 5 μg/mL concentration, and an isotypic unrelated antibody was used as negative control (Dako Denmark). A polymeric 2-step method (Supersensitive IHC Detection System, Biogenex) was used as a revelation system, according to the manufacturer's instructions, using 3,3′-diaminobenzidine as chromogen. Then, the slides were counterstained with hematoxylin, cover-slipped, and analyzed under an IX70 microscope (Olympus Biosystem) equipped with a charged coupled device camera (Camedia 4040Zoom, Olympus with a 20× objective).16
LCM and RNA extraction
Paraffin-embedded sections (8-μ thick) of LN obtained from 24 cHL patients (15 NS and 9 MC) and 15 normal LNs were fixed on PEN membrane glass slides (MDS Analytical Technologies). Additional 4-μ-thick sections were stained with hematoxylin and eosin to appreciate morphology or used for immunohistochemistry. Then tissue sections were dried at room temperature under a chemical safety hood for 5 minutes, dipped in xylene for 10 minutes twice for each sample, followed by a 3-step immersion in 100%/95%/75% ethanol solution. Samples were then washed in DEPC RNAse-free water for 1 minute, treated with a staining solution (Histogene Acturus), and dipped in 100%/95%/75% ethanol solution for 30 seconds each passage followed by xylene for 5 minutes. Tissue sections were then dried at room temperature. Some samples were processed as whole slides, and total RNA was extracted as described in the next section, whereas serial sections underwent laser capture microdissection (LCM). LCM was performed with the Veritas machine (Acturus Bioscience) to separate the stroma, identified also on the basis of TG staining, from the neoplastic fraction of each sample.32 RNA was extracted with the Paradise TM Reagent System (Acturus Bioscience) after incubation with proteinase K for 4 to 6 hours at 56°C. A DNAse treatment step was included. RNA was diluted in 50 μL elution buffer, according to the manufacturer's protocol and quantitated by NanoDrop Spectrophotometer (ND-1000 Celbio) and by Qubit TM fluorometer (Invitrogen) using the Quant-it TM Assay Kit (Invitrogen).
cDNA RT and quantitative RT-PCR
cDNA synthesis was performed with random hexamers by the use of the High Capacity Archive Kit (Applied Biosystems). To verify quantitative RT-PCR efficiency, decreasing amounts (50 ng, 10 ng, and 0.1 ng) of normal RNA were used for CT titration.
The following primers and probes were purchased from Applied Biosystems: MIC-A/B (Hs00741286_m1), ULBP1 (Hs00360941_m1), ULBP2 (Hs00607609_m1), ULBP3 (Hs00225909_m1), ULBP4 (Hs01026642_m1), NKG2D (Hs00183683_m1), ERp5 (PDIA6, Hs00194922_m1), ADAM10 (Hs00153853_m1), TG (Hs00190278_m1), IL4 (Hs00174122_m1), IL10 (Hs00961619_m1), IL-15 (Hs01003716_m1), and TGF-β Hs00998130_m1. Quantitative RT-PCR was performed on the 7900HT FastRT-PCR system (Applied Biosystems) with the fluorescent TaqMan method. mRNAs were normalized to RPLP0 as a control gene and referred to a standard curve (Ipsogen). After subtracting the threshold cycle (Ct) value for RPLP0 from the Ct values of the target genes, results were expressed as ΔCt.33,34
Statistical analysis
Data are presented as mean ± SD. Statistical analysis was performed using ANOVA for repeated measures, calculating the F ratio and, when corrected, applying the Bonferroni-Dunnett test with a α = 0.05. The cut-off value of significance was .01.
Results
Low NKG2D-L expression in cHL showing in situ high levels of ERp5 and ADAM10
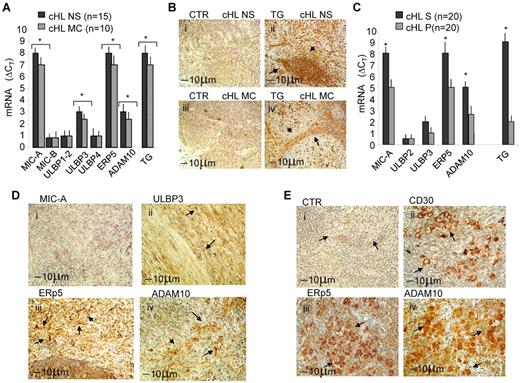
Twenty-five classic HL (cHL, 15 NS and 10 MC, Table 1) were studied for the expression of NKG2D-L. First, quantitative RT-PCR on RNA extracted from the whole LN sections, showed that MIC-A and ULBP3 transcripts were expressed in both NS and MC cHL, at variance with ULBP2, MIC-B, ULBP1, and ULBP4 (Figure 1A); these 2 NKG2D-L have recently been reported to activate γδ T cells and determine their anti–leukemia cytolytic activity.35,36 Of note, a high expression of the transcripts for both disulfide isomerase ERp5 and metalloproteinase ADAM10 could be detected in all cHLs (Figure 1A). At variance, we did not find overexpression of ADAM17 (not shown), whose catalytic domain is related to that of ADAM10, and it is also involved in NKG2D-L shedding.22 Then, molecular analysis was performed on the stromal (S) or parenchymal (P) microdissected areas of serial LN sections that underwent LCM on the basis of the localization of TG as a marker of stromal cells37 ; indeed, in both NS and MC cHL, stromal areas could be clearly detected (Figure 1Bii,iv). Quantitative RT-PCR of microdissected samples revealed that MIC-A and ULBP3 transcripts were detectable in samples derived from stromal areas, defined also by the presence of TG transcripts, at higher levels than in parenchymal fractions, both in NS and in MC cHL; likewise, ERp5 and ADAM10 enzymes were more evident in the stromal samples, although present at significant levels in the parenchyma as well (Figure 1C). Little expression, not exceeding that of housekeeping gene, of either NKG2D-L or ERp5 and ADAM10 enzymes, was detected in healthy LN (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
NKG2D-L, ERp5, and ADAM10 expression in cHL. Twenty-five cHLs were studied. (A) Quantitative RT-PCR for MIC-A/B, ULBP1 to ULBP4, TG, ERp5, and ADAM10 was performed with the specific primers and probes on RNA extracted from the whole LN sections (8 μm). After subtracting the Ct value for RPLP0 from the Ct values of the target genes, results were expressed as ΔCt. Data are the mean ± SD from 15 NS samples (black columns) and 10 MC samples (gray columns). *P < .001 versus healthy LN (supplemental Figure 1A). (B) TG staining on NS (Bii) or MC (Biv) cHL sections (4 μm) One representative staining of 20 (11 NS and 9 MC). (Bi,iii) Negative control (CTR) on NS or MC serial sections, respectively. (C) Quantitative RT-PCR of stromal (S) or parenchymal (P) microdissected areas of serial sections (n = 20 cHL: 11 NS and 9 MC) that underwent LCM on the basis of the localization of TG. Quantitative RT-PCR for MIC-A/B, ULBP2, ULBP3, TG, ERp5, and ADAM10 was performed as in panel A, normalized for RPLP0, and results calculated as ΔCt. Data are the mean ± SD from 10 S and 10 P areas cut from 20 cHL. *P < .001 S versus parenchymal. (D-E) Immunostaining for MIC-A (Di), ULBP3 (Dii), ERp5 (Diii,Eiii) ADAM10 (Div,Eiv), and CD30 (Dii) on LN sections from a representative NS (D) or MC (E) cHL of 11 NS and 9 MC analyzed. Arrows indicate positive areas. (Ei) Negative control. Arrows in panel E indicates RS cells. The slides were analyzed under an IX70 microscope (Olympus Biosystem) equipped with a camera (Camedia 4040Zoom, Olympus; original magnification ×200 with a 20×/0.50 NA objective). Black bar represents 10 μm.
NKG2D-L, ERp5, and ADAM10 expression in cHL. Twenty-five cHLs were studied. (A) Quantitative RT-PCR for MIC-A/B, ULBP1 to ULBP4, TG, ERp5, and ADAM10 was performed with the specific primers and probes on RNA extracted from the whole LN sections (8 μm). After subtracting the Ct value for RPLP0 from the Ct values of the target genes, results were expressed as ΔCt. Data are the mean ± SD from 15 NS samples (black columns) and 10 MC samples (gray columns). *P < .001 versus healthy LN (supplemental Figure 1A). (B) TG staining on NS (Bii) or MC (Biv) cHL sections (4 μm) One representative staining of 20 (11 NS and 9 MC). (Bi,iii) Negative control (CTR) on NS or MC serial sections, respectively. (C) Quantitative RT-PCR of stromal (S) or parenchymal (P) microdissected areas of serial sections (n = 20 cHL: 11 NS and 9 MC) that underwent LCM on the basis of the localization of TG. Quantitative RT-PCR for MIC-A/B, ULBP2, ULBP3, TG, ERp5, and ADAM10 was performed as in panel A, normalized for RPLP0, and results calculated as ΔCt. Data are the mean ± SD from 10 S and 10 P areas cut from 20 cHL. *P < .001 S versus parenchymal. (D-E) Immunostaining for MIC-A (Di), ULBP3 (Dii), ERp5 (Diii,Eiii) ADAM10 (Div,Eiv), and CD30 (Dii) on LN sections from a representative NS (D) or MC (E) cHL of 11 NS and 9 MC analyzed. Arrows indicate positive areas. (Ei) Negative control. Arrows in panel E indicates RS cells. The slides were analyzed under an IX70 microscope (Olympus Biosystem) equipped with a camera (Camedia 4040Zoom, Olympus; original magnification ×200 with a 20×/0.50 NA objective). Black bar represents 10 μm.
As for the expression of the proteins in situ, we found that, despite their considerable levels of transcription, MIC-A and ULBP3 were barely detectable in the LN sections of the cHL studied (Figure 1Di-ii shows a representative NS case of 11 NS and 9 MC analyzed). In turn, both the disulfide isomerase ERp5 (Figure 1Diii) and the metalloproteinase ADAM10 (Figure 1Div) were present in the stromal areas of the same LN sample. Of note, the 2 enzymes were also evident in RS cells (Figure 1Eiii-iv, respectively), identified in a representative MC case of cHL (Figure 1E) by morphologic characteristics and CD30 staining (Figure 1Eii).
ERp5+/ADAM10+ LN MSCs and RS cells can be isolated from cHL: shedding of MIC-A and ULBP3
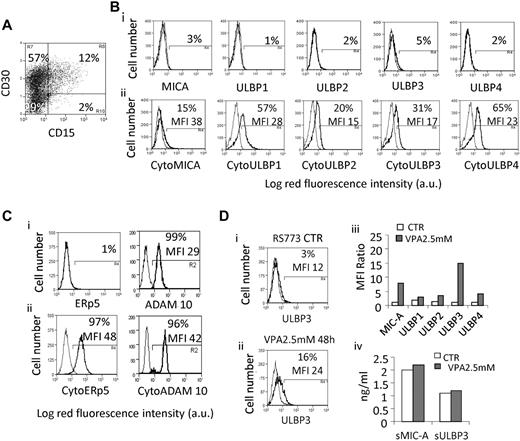
LN MSCs were obtained by culturing LN cell suspensions from cHL patients (n = 8, 6 NS, 2 MC). LN MSCs expressed HLA-I, SH3/CD73a, SH4/CD73b, SH2/CD105, PH4, collagen, vimentin, TG (Figure 2A), bone sialoprotein, osteopontin, CD44, β1-integrin/CD29, ICAM1/CD54, alkaline phosphatase, but not CD45, CD31, CD34, CD33, CD3, CD2, CD16, CD14, ICAM2, ICAM3, CD80, CD86, CD83, and HLA-DR (not shown). None of the NKG2D-L was expressed at the surface of LN MSCs, except ULBP3 (Figure 2Bi-ii), whereas all NKG2D-L could be clearly detected in the cytoplasm (Figure 2Biii-iv), where also ERp5 and ADAM10 were found at high levels (Figure 2Ciii-iv). Of note, the soluble forms of MIC-A and ULBP3 were detected in the supernatant of cultured LN MSCs (Figure 2D). In one case of cHL, we could isolate and culture also neoplastic cells with some phenotypic characteristics of RS cells. The karyotype of these cells is: 49,XY +5,+9,+12,+13 × 2, −17, −20(1). As shown in Figure 3A, these cells were mostly CD30 (70%), and a fraction coexpressed the CD15 molecules, as described for RS cells.37 The NKG2D-L ligands were not detectable at the surface of RS773 cells by immunofluorescence (Figure 3Bi), and ULBPs 1 to 4 were retained in the cytoplasm (Figure 3Bii); on the other hand, they showed cytoplasmic ERp5 and ADAM10 (Figure 3Cii), the latter also detectable at the cell surface (Figure 3Ci). These cells were kept in culture as a cell line named RS773 and used for further experiments. Because NKG2D-L surface expression can be induced by ATRA8,15,27 or VPA,27,38,39 we checked MIC-A and ULBPs 1 to 4 by immunofluorescence after in vitro exposure of RS773 cells to 10μM ATRA or 2.5mM VPA. As shown in Figure 3D, 2.5mM VPA could induce MIC-A (Figure 3Diii) and ULBP3 (Figure Dii-iii) at the surface of RS773, at variance with ATRA that did not exert any effect on this cell line (not shown). Cultured RS773 could release both sMIC-A and sULBP3; the shedding of these molecules was not significantly affected by treatment with VPA (Figure 3Div).
Expression of NKG2D-L, ERp5, and ADAM10 in LN MSCs. LN MSCs were obtained from cHL patients (n = 8, 6 NS, 2 MC). (A) Staining with specific mAbs recognizing HLA-I, SH3/CD73a, CD90, SH2/CD105 (Ai), PH4, collagen, vimentin, and TG (Aiii), followed by PE-conjugated anti–isotype specific GAM (black lines). Gray lines indicate negative control staining with isotype-matched irrelevant mAbs followed by PE-GAM. Samples were run on a CyAnADP flow cytometer, and results are expressed as log red MFI (a.u. indicates arbitrary units) versus number of cells; in each histogram are indicated the MFI and the percentage of positive cells. (Aii,iv) MFI ratio between MFI of positive cells for each marker and MFI of the negative control. Data are mean ± SD from 8 LN MSCs. (B) Surface (Bi) or cytoplasmic (Biii) staining of LN MSCs with anti–MIC-A or anti-ULBP1 to ULBP4 mAbs. For cytoplasmic staining, cells were fixed in 3% paraformaldehyde and permeabilized with1% Nonidet-P40 in PBS, before mAb incubation. Results are expressed as mean red fluorescence intensity; in each histogram are shown the MFI and the percentage of positive cells. (Bii,iv) MFI ratio as in panel A. Data are mean ± SD from 8 LN MSCs. (C) Surface (Ci) and cytoplasmic (Ciii) staining of LN MSCs with the anti–ERp5 rabbit polyclonal antibody or anti–ADAM10 mAb, followed by FITC-goat anti–rabbit (GAR) or anti–isotype GAM antisera (black lines). Gray lines indicate negative control staining with unrelated irrelevant Abs. Results are expressed as log green mean fluorescence intensity; in each histogram are indicated the MFI and the percentage of positive cells. (Cii,iv) MFI ratio: ratio between MFI of a given marker and the negative control. Data are mean ± SD from 8 LN MSCs. (D) Soluble MIC-A, ULBP2, and ULBP3 were measured in the supernatant of cultured LN MSCs by ELISA, referred to a standard curve, and expressed as nanograms per milliliter. Data are mean ± SD from 8 LN MSCs.
Expression of NKG2D-L, ERp5, and ADAM10 in LN MSCs. LN MSCs were obtained from cHL patients (n = 8, 6 NS, 2 MC). (A) Staining with specific mAbs recognizing HLA-I, SH3/CD73a, CD90, SH2/CD105 (Ai), PH4, collagen, vimentin, and TG (Aiii), followed by PE-conjugated anti–isotype specific GAM (black lines). Gray lines indicate negative control staining with isotype-matched irrelevant mAbs followed by PE-GAM. Samples were run on a CyAnADP flow cytometer, and results are expressed as log red MFI (a.u. indicates arbitrary units) versus number of cells; in each histogram are indicated the MFI and the percentage of positive cells. (Aii,iv) MFI ratio between MFI of positive cells for each marker and MFI of the negative control. Data are mean ± SD from 8 LN MSCs. (B) Surface (Bi) or cytoplasmic (Biii) staining of LN MSCs with anti–MIC-A or anti-ULBP1 to ULBP4 mAbs. For cytoplasmic staining, cells were fixed in 3% paraformaldehyde and permeabilized with1% Nonidet-P40 in PBS, before mAb incubation. Results are expressed as mean red fluorescence intensity; in each histogram are shown the MFI and the percentage of positive cells. (Bii,iv) MFI ratio as in panel A. Data are mean ± SD from 8 LN MSCs. (C) Surface (Ci) and cytoplasmic (Ciii) staining of LN MSCs with the anti–ERp5 rabbit polyclonal antibody or anti–ADAM10 mAb, followed by FITC-goat anti–rabbit (GAR) or anti–isotype GAM antisera (black lines). Gray lines indicate negative control staining with unrelated irrelevant Abs. Results are expressed as log green mean fluorescence intensity; in each histogram are indicated the MFI and the percentage of positive cells. (Cii,iv) MFI ratio: ratio between MFI of a given marker and the negative control. Data are mean ± SD from 8 LN MSCs. (D) Soluble MIC-A, ULBP2, and ULBP3 were measured in the supernatant of cultured LN MSCs by ELISA, referred to a standard curve, and expressed as nanograms per milliliter. Data are mean ± SD from 8 LN MSCs.
NKG2D-L, ERp5, and ADAM10 expression in the RS773 cell line. (A) Double staining of the RS773 cell line, derived from one case of MC cHL, with PE–anti-CD30 and FITC–anti-CD15 mAbs. Samples were run on a CyAnADP flow cytometer, and results are expressed as log red fluorescence intensity versus log green fluorescence intensity (percentages of single- or double-positive cells in each quadrant). Quadrants were set on the negative control stained with unrelated mAb (not shown). (B) Surface (Bi) or cytoplasmic (Bii) staining of RS773 for MIC-A, ULBP1, ULBP2, ULBP3, and ULBP4, with the specific mAbs followed by PE-GAM. For cytoplasmic staining, cells were fixed in 3% paraformaldehyde and permeabilized with1% Nonidet-P40 in PBS, before mAb incubation. Samples were run on a CyAnADP flow cytometer, and results are expressed as log MFI (a.u.) versus number of cells. (C) Surface (Ci) or cytoplasmic (Cii) staining of RS773 cells for ERp5 and ADAM10, with the rabbit polyclonal anti-ERp5 antiserum or the anti–ADAM10 mAb, followed by FITC-GAR or anti–isotype GAM antisera. For cytoplasmic staining, cells were fixed and permeabilized as in panel B, before Ab incubation. Results are shown as overlay histograms between the negative control (thin gray line) and the sample (black line). (D) MIC-A and ULBP1 to ULBP4 surface staining performed as in panel B, before (Di,iii) or after in vitro exposure of RS773 cells to 2.5mM VPA for 48 hours (Dii,iii). Results are expressed as log MFI (a.u.) versus number of cells (Di,ii) or as MFI ratio (ie, MFI of the sample/MFI of the negative control; Diii). (Div) Soluble MIC-A and ULBP3 were measured in the supernatants of cultured RS773, either untreated (CTR) or treated with 2.5mM VPA, by ELISA, referred to a standard curve and expressed as nanograms per milliliter.
NKG2D-L, ERp5, and ADAM10 expression in the RS773 cell line. (A) Double staining of the RS773 cell line, derived from one case of MC cHL, with PE–anti-CD30 and FITC–anti-CD15 mAbs. Samples were run on a CyAnADP flow cytometer, and results are expressed as log red fluorescence intensity versus log green fluorescence intensity (percentages of single- or double-positive cells in each quadrant). Quadrants were set on the negative control stained with unrelated mAb (not shown). (B) Surface (Bi) or cytoplasmic (Bii) staining of RS773 for MIC-A, ULBP1, ULBP2, ULBP3, and ULBP4, with the specific mAbs followed by PE-GAM. For cytoplasmic staining, cells were fixed in 3% paraformaldehyde and permeabilized with1% Nonidet-P40 in PBS, before mAb incubation. Samples were run on a CyAnADP flow cytometer, and results are expressed as log MFI (a.u.) versus number of cells. (C) Surface (Ci) or cytoplasmic (Cii) staining of RS773 cells for ERp5 and ADAM10, with the rabbit polyclonal anti-ERp5 antiserum or the anti–ADAM10 mAb, followed by FITC-GAR or anti–isotype GAM antisera. For cytoplasmic staining, cells were fixed and permeabilized as in panel B, before Ab incubation. Results are shown as overlay histograms between the negative control (thin gray line) and the sample (black line). (D) MIC-A and ULBP1 to ULBP4 surface staining performed as in panel B, before (Di,iii) or after in vitro exposure of RS773 cells to 2.5mM VPA for 48 hours (Dii,iii). Results are expressed as log MFI (a.u.) versus number of cells (Di,ii) or as MFI ratio (ie, MFI of the sample/MFI of the negative control; Diii). (Div) Soluble MIC-A and ULBP3 were measured in the supernatants of cultured RS773, either untreated (CTR) or treated with 2.5mM VPA, by ELISA, referred to a standard curve and expressed as nanograms per milliliter.
Taken together, these results indicate that the low expression of transmembrane NKG2D-L and high levels of their soluble form may be the result of the presence of the disulfide isomerase ERp5 and the disintegrin-metalloproinase ADAM10 in LN MSCs and/or RS cells.
Impaired recognition of lymphoma cells: effect of coculture of T lymphocytes with LN MSCs
We next investigated the role of LN MSCs in the modulation of lymphoma cell recognition by effector T lymphocytes. To this aim, CD8+αβT or γδT lymphocytes, either Vδ1 or Vδ2, were used as effector cells against RS773, L-540, KM-H2, L-428, and HDLM-2 Hodgkin cell lines in a cytotoxicity assay. First, CD8+αβ or γδT lymphocyte-mediated killing of RS773 cells that do not bear NKG2D-L (Figure 3B) and do not bind the NKG2D soluble receptor (NKG2DFc, Figure 4Ai) is low even at high E:T ratios (10:1, Figure 4Aii), undetectable at lower E:T ratio as 5:1 or 2:1 (not shown); similar results were obtained with the KM-H2 cell line (Figure 4Bii) that expresses very low levels of ULBP3 (supplemental Figure 2A) and a barely detectable binding of the NKG2D Fc soluble receptor (Figure 4Bi). A higher degree of cytotoxicity was observed when RS773, or the KM-H2 cell line, pretreated with VPA, thus expressing ULBPs and binding the NKG2D receptor (Figure 4Ci,Di), was used as a target (Figure 4Cii,Dii). In all cases, blocking of NKG2D at the surface of T cells with a specific mAb strongly reduced their cytolytic activity (Figure 4Aii-Dii). Interestingly, on coculture with LN MSCs, CD8+ T lymphocytes strongly reduced their capability of killing either RS773 or KM-H2 cell lines (Figure 4E-F), even if the targets underwent VPA treatment, thus increasing NKG2D-L expression (Figure 3Diii; supplemental Figure 2A-B) and reactivity with the NKG2D receptor (Figure 4Ci vs Figure 4Ai; and Figure 4Di vs Figure 4Bi); this inhibitory effect exerted by LN MSCs was only partially prevented when cocultures were performed in the presence of the soluble NKG2D Fc, to neutralize the sMIC-A and/or sULBP3 released by LN MSC, or with an anti–TGF-β mAb to prevent down-regulation of NKG2D expression (Figures 4E-F and 5). Similar results were obtained with all the cHL cell lines or using γδ T lymphocytes as effector cells (not shown). Supplemental Figure 2 shows induction of NKG2D ligands by 2.5mM VPA (supplemental Figure 2A) and the cytoplasmic expression of ERp5 and ADAM 10 (supplemental Figure 2C) on KM-H2 cell line. Supplemental Figure 2B,Cii shows the same results as the mean ± SD from the 4 cHL cell lines.
Effect of T lymphocyte-LN MSC coculture on anti–lymphoma cytotoxicity. CD8+αβT or γδT lymphocytes, Vδ1 or Vδ2 as indicated, were used as effector cells, whereas the RS773 (A,C,E) and the KM-H2 Hodgkin RS-like cell lines (B,D,F), either untreated or pretreated for 48 hours with 2.5mM VPA (C-F), were used as targets in a 4-hour 51Cr-release assay at the E:T ratios of 10:1. In some experiments, the effector T lymphocytes were precultured on LN MSCs for 5 days before using in cytolytic assay. In some experiments (E-F), the soluble NKG2D receptor (NKG2D-Fc) or the anti–TGF-β mAb (5 μg/mL) was added to lymphocyte-LN MSCs cocultures; then effector cells washed and used in cytolytic assay. In all cytotoxicity experiments, a blocking anti–NKG2D specific mAb (5 μg/mL) was used compared with an unrelated, isotype-matched mAb. (A-D) Subpanel i shows the reactivity of the soluble NKG2D receptor (NKG2D Fc), evidenced with a PE-goat anti–human, to the RS773 or KM-H2 cell lines before (Ai,Bi) or after VPA exposure (Ci,Di). Results are expressed as percentage of specific cell lysis and are the mean ± SD from 6 experiments performed with 6 different LN MSCs. (Cii,Dii) *P < .001 versus RS773 in panel Aii and versus KM-H2 in panel Bii. **P < .001 versus cytolytic activity of effector cells in the absence of anti–NKG2D mAb. (E-F) *P < .001 versus cell lysis exerted by CD8+ T lymphocytes before LN MSC coculture and/or in the absence of anti–NKG2D mAb. **P < .001 versus cytolytic activity exerted by CD8+ T lymphocytes after LN MSC coculture.
Effect of T lymphocyte-LN MSC coculture on anti–lymphoma cytotoxicity. CD8+αβT or γδT lymphocytes, Vδ1 or Vδ2 as indicated, were used as effector cells, whereas the RS773 (A,C,E) and the KM-H2 Hodgkin RS-like cell lines (B,D,F), either untreated or pretreated for 48 hours with 2.5mM VPA (C-F), were used as targets in a 4-hour 51Cr-release assay at the E:T ratios of 10:1. In some experiments, the effector T lymphocytes were precultured on LN MSCs for 5 days before using in cytolytic assay. In some experiments (E-F), the soluble NKG2D receptor (NKG2D-Fc) or the anti–TGF-β mAb (5 μg/mL) was added to lymphocyte-LN MSCs cocultures; then effector cells washed and used in cytolytic assay. In all cytotoxicity experiments, a blocking anti–NKG2D specific mAb (5 μg/mL) was used compared with an unrelated, isotype-matched mAb. (A-D) Subpanel i shows the reactivity of the soluble NKG2D receptor (NKG2D Fc), evidenced with a PE-goat anti–human, to the RS773 or KM-H2 cell lines before (Ai,Bi) or after VPA exposure (Ci,Di). Results are expressed as percentage of specific cell lysis and are the mean ± SD from 6 experiments performed with 6 different LN MSCs. (Cii,Dii) *P < .001 versus RS773 in panel Aii and versus KM-H2 in panel Bii. **P < .001 versus cytolytic activity of effector cells in the absence of anti–NKG2D mAb. (E-F) *P < .001 versus cell lysis exerted by CD8+ T lymphocytes before LN MSC coculture and/or in the absence of anti–NKG2D mAb. **P < .001 versus cytolytic activity exerted by CD8+ T lymphocytes after LN MSC coculture.
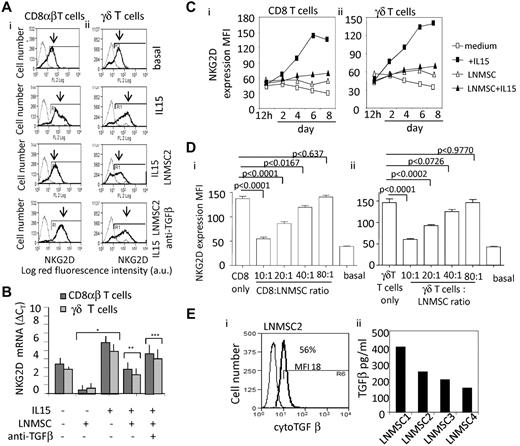
Down-regulation of NKG2D on effector cells by TGF-β produced by LN MSCs from HL. NKG2D expression was evaluated by indirect immunofluorescence on coculture of CD8+αβ (Ai,Ci,Di) and γδ T lymphocytes (Aii,Cii,Dii) with LN MSCs, in the presence of IL-15 (10 ng/mL) and in the absence or presence of an anti–TGF-β mAb (5 μg/mL) added to the cocultures (A). NKG2D was evidenced by a specific mAb followed by PE-GAM. Samples were run on a CyAnADP flow cytometer, and results are expressed as log MFI (a.u.) versus number of cells (A) or as MFI (a.u., C-D). Results are shown as overlay histograms of negative control (gray thin line) and NKG2D mAb reactivity (black thick line, as indicated by the arrows). (B) Quantitative RT-PCR for NKG2D was performed with the specific primers and probes on RNA extracted from CD8+ αβ (dark gray columns) and γδ T cells (light gray columns), after 24 hours or culture without or with IL-15 (10 ng/mL) and/or anti–TGF-β mAb (5 μg/mL), in the presence or absence of LN MSCs, as indicated. Data are mean ± SD from 3 experiments with CD8+ αβ T cells and 3 with γδ T cells. *P < .001versus experiments done in the absence of LN MSCs and IL-15. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. (C) Kinetics of NKG2D intensity of expression (MFI, a.u.) in CD8+ αβ (Ci) and γδT lymphocytes (Cii) cocultured with LN MSCs in the presence of IL-15 or in medium alone (medium). (D) NKG2D intensity of expression (MFI, a.u.) in CD8+ αβ (Di) and γδ T lymphocytes (Dii) cocultured with LN MSCs at different ratios (as indicated) in the presence of IL-15 or in medium alone (basal). Data are mean ± SD from 6 experiments performed with 6 different LN MSCs. (E) TGF-β in the cytoplasm (Ei, 1 representative experiment) and in the supernatant of cultured LN MSCs (Eii, 4 different LN MSCs). (Ei) For cytoplasmic staining, cells were fixed and permeabilized before addition of anti–TGF-β mAb, followed by FITC-GAM. Samples were run on CyAnADP flow cytometer, and results are expressed as MFI versus number of cells. (Eii) TGF-β1 was measured in SN recovered from LN MSC cultures after 24 hours, on treatment for 1 hour of each SN with 1N HCl followed by 1N NaOH, with a TGF-β1 specific ELISA kit. Results were normalized to a standard curve and expressed as picograms per milliliter.
Down-regulation of NKG2D on effector cells by TGF-β produced by LN MSCs from HL. NKG2D expression was evaluated by indirect immunofluorescence on coculture of CD8+αβ (Ai,Ci,Di) and γδ T lymphocytes (Aii,Cii,Dii) with LN MSCs, in the presence of IL-15 (10 ng/mL) and in the absence or presence of an anti–TGF-β mAb (5 μg/mL) added to the cocultures (A). NKG2D was evidenced by a specific mAb followed by PE-GAM. Samples were run on a CyAnADP flow cytometer, and results are expressed as log MFI (a.u.) versus number of cells (A) or as MFI (a.u., C-D). Results are shown as overlay histograms of negative control (gray thin line) and NKG2D mAb reactivity (black thick line, as indicated by the arrows). (B) Quantitative RT-PCR for NKG2D was performed with the specific primers and probes on RNA extracted from CD8+ αβ (dark gray columns) and γδ T cells (light gray columns), after 24 hours or culture without or with IL-15 (10 ng/mL) and/or anti–TGF-β mAb (5 μg/mL), in the presence or absence of LN MSCs, as indicated. Data are mean ± SD from 3 experiments with CD8+ αβ T cells and 3 with γδ T cells. *P < .001versus experiments done in the absence of LN MSCs and IL-15. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. (C) Kinetics of NKG2D intensity of expression (MFI, a.u.) in CD8+ αβ (Ci) and γδT lymphocytes (Cii) cocultured with LN MSCs in the presence of IL-15 or in medium alone (medium). (D) NKG2D intensity of expression (MFI, a.u.) in CD8+ αβ (Di) and γδ T lymphocytes (Dii) cocultured with LN MSCs at different ratios (as indicated) in the presence of IL-15 or in medium alone (basal). Data are mean ± SD from 6 experiments performed with 6 different LN MSCs. (E) TGF-β in the cytoplasm (Ei, 1 representative experiment) and in the supernatant of cultured LN MSCs (Eii, 4 different LN MSCs). (Ei) For cytoplasmic staining, cells were fixed and permeabilized before addition of anti–TGF-β mAb, followed by FITC-GAM. Samples were run on CyAnADP flow cytometer, and results are expressed as MFI versus number of cells. (Eii) TGF-β1 was measured in SN recovered from LN MSC cultures after 24 hours, on treatment for 1 hour of each SN with 1N HCl followed by 1N NaOH, with a TGF-β1 specific ELISA kit. Results were normalized to a standard curve and expressed as picograms per milliliter.
TGF-β produced by LN MSCs from HL down-regulates NKG2D expression on effector cells
Because TGF-β is known to down-regulate the expression of NKG2D receptor at the cell surface,24-26 we addressed the question of whether this might occur also in the microenvironment of cHL, thus contributing, together with the release of soluble NKG2D-L, to the impairment of lymphoma cell killing. Interestingly, the up-regulation of NKG2D expression induced by IL-15 in vitro was strongly inhibited by coculture of CD8+ αβ and CD8+ γδ T lymphocytes with LN MSCs (Figure 5Ai-ii); this down-regulation was observed at 24 hours as mRNA (Figure 5B) and, starting from day 3 of culture at the protein level (Figure 5Ci-ii), was dependent on the T:LN MSC ratio (Figure 5Di-ii), mimicked by exposure of T cells to purified TGF-β (not shown) and prevented by an anti–TFG-β mAb added to the cocultures (Figure 5Ai-ii,B). Along this line, TGF-β was detected in the cytoplasm of cultured LN MSCs (Figure 5Ei) and measured in their supernatants (Figure 5Eii).
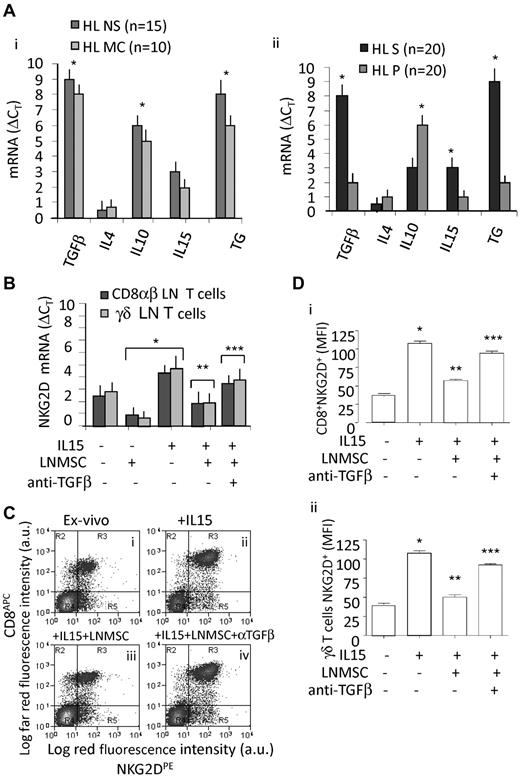
Of note, TGF-β was transcribed at high levels at the tumor site in all cHL tested, mainly in the stromal microdissected areas, where also IL-15 transcripts were detectable (Figure 6Ai for the whole sample and Aii for microdissected sections); in turn, the parenchymal areas were enriched in IL-10 transcripts (Figure 6Aii). No expression of IL-4, which we described in NHL,16 was found in cHL (Figure 6Ai-ii). Then, lymphocytes isolated from cHL LN specimens were evaluated for NKG2D transcription (by quantitative RT-PCR; Figure 6B) or expression by indirect immunofluorescence (Figure 6C-D), either freshly isolated (Figure 6Ci) or cultured for 6 days with IL-15 (Figure 6Cii) or on coculture with LN MSCs, in the presence of IL-15 (Figure 6Ciii) and in the presence of IL-15 and of an anti–TFG-β mAb (Figure 6Civ). Figure 6D shows NKG2D intensity of expression (MFI arbitrary units) in CD8+ αβ (Figure 6Di) and γδ T lymphocytes (Figure 6Dii) cocultured with LN MSCs in the absence or presence of IL-15 or of an anti–TFG-β mAb as the mean ± SD from 6 experiments performed with 6 different LN MSCs.
TGF-β is expressed in the cHL microenvironment. (A) Quantitative RT-PCR for TGF-β, IL-4, IL-10, and TG was performed with the specific primers and probes on RNA extracted from the whole LN sections (8 μm, Ai) or on stromal (S) or parenchymal (P) microdissected areas of serial sections (n = 20 cHL: 11 NS and 9 MC) that underwent LCM on the basis of the localization of TG (Aii). After subtracting the Ct value for RPLP0 from the Ct values of the target genes, results were expressed as ΔCt. Data are the mean ± SD from 14 NS samples (black columns) and 9 MC samples (gray columns in Ai). *P < .001 versus healthy LN (supplemental Figure 1A). (B) Quantitative RT-PCR for NKG2D was performed with the specific primers and probes on RNA extracted from CD8+ αβ (dark gray columns) and γδ T cells (light gray columns) isolated from LN, after 24 hours or culture without or with IL-15 (10 ng/mL), in the presence or absence of LN MSCs, anti–TGF-β mAb (5 μg/mL), as indicated. Data are mean ± SD from 3 experiments with CD8+ αβ and 3 with γδ T cells. *P < .001versus experiments done in the absence of LN MSCs and IL-15. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. (C) Lymphocytes isolated from 6 HL LN specimens were evaluated for NKG2D expression by indirect immunofluorescence, either freshly isolated (Ci) or cultured for 6 days with rIL-15 (10 ng/mL, Bii) or on coculture with LN MSCs, in the presence of IL-15 (Ciii) and in the presence of IL-15 and of an anti–TFG-β mAb (5 μg/mL) added to the cocultures (Civ). NKG2D was evidenced by a specific mAb followed by PE-GAM, whereas CD8+ cells were identified by an APC-conjugated anti–CD8 mAb (C-Di), whereas γδT cells were detected with a combination of anti-Vδ1 and anti-Vδ2 AlexaFluor-647–conjugated mAbs (Dii). Samples were run on a CyAnADP flow cytometer, and results are expressed as log mean red fluorescence intensity (x-axis MFI, a.u.) versus log mean far red fluorescence intensity (for both APC and AlexaFluor-647, y-axis MFI, a.u.), or as MFI (a.u.). (D) Data are mean ± SD from 6 different experiments with CD8+ cells (Di) or γδ T cells (Dii) and LN MSCs from 6 different HLs. *P < .001 versus experiments done in the absence of IL-15, LN MSCs, or anti–TGF-β. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. All results were determined on CD3+ gated T cells after simultaneous staining with anti-CD3 mAb (JT3A, IgG2a) and FITC-GAM.
TGF-β is expressed in the cHL microenvironment. (A) Quantitative RT-PCR for TGF-β, IL-4, IL-10, and TG was performed with the specific primers and probes on RNA extracted from the whole LN sections (8 μm, Ai) or on stromal (S) or parenchymal (P) microdissected areas of serial sections (n = 20 cHL: 11 NS and 9 MC) that underwent LCM on the basis of the localization of TG (Aii). After subtracting the Ct value for RPLP0 from the Ct values of the target genes, results were expressed as ΔCt. Data are the mean ± SD from 14 NS samples (black columns) and 9 MC samples (gray columns in Ai). *P < .001 versus healthy LN (supplemental Figure 1A). (B) Quantitative RT-PCR for NKG2D was performed with the specific primers and probes on RNA extracted from CD8+ αβ (dark gray columns) and γδ T cells (light gray columns) isolated from LN, after 24 hours or culture without or with IL-15 (10 ng/mL), in the presence or absence of LN MSCs, anti–TGF-β mAb (5 μg/mL), as indicated. Data are mean ± SD from 3 experiments with CD8+ αβ and 3 with γδ T cells. *P < .001versus experiments done in the absence of LN MSCs and IL-15. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. (C) Lymphocytes isolated from 6 HL LN specimens were evaluated for NKG2D expression by indirect immunofluorescence, either freshly isolated (Ci) or cultured for 6 days with rIL-15 (10 ng/mL, Bii) or on coculture with LN MSCs, in the presence of IL-15 (Ciii) and in the presence of IL-15 and of an anti–TFG-β mAb (5 μg/mL) added to the cocultures (Civ). NKG2D was evidenced by a specific mAb followed by PE-GAM, whereas CD8+ cells were identified by an APC-conjugated anti–CD8 mAb (C-Di), whereas γδT cells were detected with a combination of anti-Vδ1 and anti-Vδ2 AlexaFluor-647–conjugated mAbs (Dii). Samples were run on a CyAnADP flow cytometer, and results are expressed as log mean red fluorescence intensity (x-axis MFI, a.u.) versus log mean far red fluorescence intensity (for both APC and AlexaFluor-647, y-axis MFI, a.u.), or as MFI (a.u.). (D) Data are mean ± SD from 6 different experiments with CD8+ cells (Di) or γδ T cells (Dii) and LN MSCs from 6 different HLs. *P < .001 versus experiments done in the absence of IL-15, LN MSCs, or anti–TGF-β. **P < .001 versus experiments in the presence of IL-15. ***P < .001 versus experiments in the presence of IL-15 and LN MSCs. All results were determined on CD3+ gated T cells after simultaneous staining with anti-CD3 mAb (JT3A, IgG2a) and FITC-GAM.
We found that NKG2D was up-regulated in culture by IL-15 on CD8+ T lymphocytes, identified by an APC-conjugated anti–CD8 mAb, and this up-regulation was prevented when the anti–TFG-β mAb was added to the cultures (Figure 6Civ,Di). Similar results were obtained analyzing NKG2D expression on Vδ1 (Figure 6Dii) or Vδ2 (not shown) T cells from the same tissue samples. It is of interest that the intensity of NKG2D expression, measured as MFI arbitrary units, was higher in CD8+ T lymphocytes isolated from 3 healthy LN (MFI: 58 ± 5 arbitrary units), than in CD8+ T cells from cHL LN (MFI: 41 ± 6 arbitrary units), thus further supporting that NKG2D receptor may be down-regulated in the cHL microenvironment.
Discussion
It is now accepted that ectopic or enhanced expression of NKG2D-L on cancer cells renders them susceptible to an anti–tumor immune response in vitro and in vivo.1-5 In hematologic malignancies, there are several evidences supporting that both αβ and γδ T cells act as anti–leukemic effectors using the NKG2D-NKG2D-L recognition.2,12-16 Nevertheless, in cHL, the infiltrating T-cell population seems to be committed toward a regulatory, rather than an effector, function.40
In this paper, we describe that in cHL the microenvironment may contribute to tumor escape from the immune system in 2 ways: (1) through a reduction of NKG2D-L expression at the surface of lymphoma cell, together with enhanced release of their soluble form; and (2) by the mean of cytokines able to down-regulate NKG2D receptor on effector lymphocytes and impair tumor cell recognition. Indeed, in all cHL analyzed, the LN stroma displayed, in situ, high levels of transcription of the disulfide isomerase ERp5 and of the disintegrin-metalloproteinase ADAM10. These enzymes, which are able to shed all NKG2D ligands from the cell membrane,20-22 were also detected as proteins, both in the stromal areas and in RS tumor cells in the LN of cHL patients and were apparently active in cultured LN MSCs, as well as in RS cells, as soluble MIC-A and ULBP3 were present in culture supernatants, but missing or expressed at very low levels on the cell surface. It is to be noted that sNKG2D-L interfere with the binding of lymphocyte NKG2D receptor to their membrane-bound form on tumor cells; moreover, RS cells (RS773 cell line), and all the cHL cell lines tested, which are negative for NKG2D-L, could not be recognized and killed by CD8+ αβ and γδ T cells. Interestingly, tumor cell recognition and killing were partially restored by treating RS773 cells, or the other cHL cell lines, with VPA that enhanced NKG2D-L transcription, conceivably to levels high enough to overcome the activity of ERp5 or ADAM10; indeed, both MIC-A and ULBP3 were detectable at the surface of RS773 cells for several days after VPA exposure. However, although not increased, the shedding of their soluble form was still observed after VPA treatment; this would imply that induction of NKG2D-L at the surface of target cells is not sufficient to maintain tumor cell killing.
Of note, on coculture with LN MSCs, both CD8+ αβ and γδ T cells strongly reduced their cytolytic activity even against NKG2D-L bearing targets; this seems to be mainly the result of TGF-β, produced and released in vitro by LN MSCs, and able to down-regulate the expression of NKG2D on effector lymphocytes.24-26 This was also supported by the finding that an anti–TGF-β mAb could prevent the inhibition of NKG2D expression and of effector cell function induced by LN MSC cocultures; in turn, both the inhibitory effect of cytotoxicity and the down-regulation of NKG2D expression by effector T lymphocytes could be reproduced using either SN from the cocultures or recombinant TGF-β (not shown). This cytokine was also detected at the tumor site, at variance with IL-4, that we described in NHL,16 and together with IL-10, in keeping with other reports.40-42 In addition, we also found that CD8+αβT and γδT lymphocytes in cell suspensions isolated from cHL undergo NKG2D up-regulation in vitro in the presence of IL-15, which is prevented by coculture with LN MSCs.
Thus, in cHL microenvironment, different molecular events may take place, involving stromal or tumor cells and infiltrating lymphocytes, all of them resulting in the failure of cancer cell recognition by immune effector cells. On one hand, high expression of ERp5 and ADAM10 by LN MSCs or RS cells, would contribute to the low availability of membrane-bound NKG2D-L in favor of their released soluble form. On the other hand, immunosuppressive cytokines, such as TGF-β and IL-10, produced by stromal cells and infiltrating lymphocytes, might lead to down-regulation of NKG2D receptor, impaired NKG2D-L binding, and tumor cell killing.
All these findings may have both prognostic and therapeutic implications. Indeed, plasma levels of sNKG2D-L correlate with disease progression in different hematologic malignancies, such as in multiple myeloma, CLL, NHL, and acute myeloid leukemias; in particular, among sNKG2D-L, both sMIC-A and sULBP2 have been shown as a prognostic marker for multiple myeloma and for the identification of early-stage CLL patients with risk of disease progression.14-16,21,27,28 In turn, expression of membrane-bound NKG2D-L by tumor cells might be up-regulated by drugs, such as ATRA or VPA15,27,31 or, as recently reported, by proteasome inhibitors,43 all already approved for the treatment of hematologic malignancies. In addition, a bispecific ULBP2-BB4 protein targeting NKG2D on cytotoxic lymphocytes and CD138 on myeloma cells has been recently shown to prevent tumor cell growth in nude mice.44 Enhancement of NKG2D-mediated anti–tumor immune response by RNA interference targeting TGF-β has been reported to inhibit tumorigenicity in vivo in an animal model.45 It of interest that also prevention of NKG2D-L release is now proposed as a potential therapeutic target: these therapies would include blocking of ERp5-binding domain on MIC-A, or inhibition of ERp5 or ADAM10 enzymatic activity.46-48
In conclusion, selective modulation of NKG2D receptor expression and control of the enzymatic regulation of the balance between soluble and membrane-bound NKG2D-L may provide an additional opportunity to potentiate anticancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Amgen (MTA no. 200309766-001) for kindly providing the anti-ULBP antibodies.
This work was supported in part by the Italian Association for Cancer Research (grants IG8761 and IG8727, A.P. and M.R.Z.) and the Compagnia di San Paolo (grants 2007.2065 and 2010.14038).
Authorship
Contribution: S.C., A.K., A.M., S.B., and P.C. performed flow cytometry, cell cultures, and molecular biology; A.Z. characterized the tumor cell lines; S.T. performed histochemistry; A.G. and B.V. performed microdissection; G.F.-O., J.-L.R., M.G., M.R.Z., and A.P. analyzed clinical and experimental data; and A.P. and M.R.Z. designed research, analyzed data, wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alessandro Poggi, Unit of Molecular Oncology and Angiogenesis, IRCCS AOU San Martino–IST National Institute for Cancer Research, L.go R.Benzi 10, I-16132 Genoa, Italy; e-mail: alessandro.poggi@istge.it; or Maria Raffaella Zocchi, Division of Immunology, Transplants, Infectious Diseases, Scientific Institute San Raffaele,Via Olgettina 60, I-20132 Milan, Italy; e-mail: zocchi.maria@hsr.it.