Abstract
T-cell immunity is important for controlling Kaposi sarcoma–associated herpesvirus (KSHV) diseases such as the endothelial cell malignancy Kaposi sarcoma, or the B-cell malignancy, primary effusion lymphoma (PEL). However, little is known about KSHV-specific T-cell immunity in healthy donors and immune control of disease. Using PBMCs from healthy KSHV-infected donors, we found weak ex vivo responses to the KSHV latent antigens LANA, vFLIP, vCyclin, and Kaposin, with LANA most frequently recognized. CD4+ T-cell clones specific to LANA, a protein expressed in all KSHV-infected cells and malignancies, were established to determine whether they could recognize LANA-expressing cells. B-cell targets expressing or fed LANA protein were consistently recognized by the clones; however, most PEL cell lines were not. PELs express the KSHV protein vIRF3 that inhibits promoter function of the HLA class II transactivator, decreasing expression of genes controlled by this transactivator. Re-expressing the class II transactivator in the PELs increased expression of downstream targets such as HLA class II and restored recognition but not killing by the LANA-specific clones. We suggest that PELs are poorly controlled in vivo because of inefficient recognition and killing by T cells.
Introduction
Kaposi sarcoma–associated herpesvirus (KSHV) is a lymphotropic human herpesvirus with oncogenic potential. KSHV infects endothelial and B cells where it can cause malignancies of these cell types, namely, Kaposi sarcoma (KS) and primary effusion lymphoma (PEL), respectively,1 and it is associated with the B-cell pathology multicentric Castleman disease (MCD). The HIV epidemic has made these diseases significantly more prevalent, with KS being the most frequently reported HIV-associated malignancy.
An important finding from studying KS patients either with HIV, or those immunosuppressed after solid organ transplantation, is that this malignancy can resolve on restoration of immune function through the administration of highly active antiretroviral therapy (HAART)2 or relaxation of immunosuppression,3 respectively. These findings and the increased incidence of PEL and MCD in HIV patients4 suggest that T-cell immunity is critical for control of KSHV infection and disease. Potential immune targets in KS include the latent antigens, namely, the genome maintenance protein LANA that is expressed in all infected cells and malignancies, the viral cyclin vCyclin, the antiapoptotic multifunctional protein vFLIP, and Kaposin. PELs and infected cells in MCD also express at least 2 other proteins, the viral IL-6 and the immunomodulatory and antiapoptotic protein vIRF3.
Relatively low T-cell responses to LANA and Kaposin have been described in healthy immunocompetent donors;5,6 however, most studies have been undertaken using donors with a disrupted immune system or in disease settings, focusing on responses to LANA and Kaposin. Thus, HIV-infected patients on HAART who control KSHV have been found to make T-cell responses to LANA, MCD patients make responses comparable to HIV patients on HAART, but patients with KS disease have very weak or no detectable responses.7–11 The administration of HAART to HIV-associated KS patients has been associated with the detection and increase in KSHV-specific responses over time.2,12 Little is known, however, about the size of responses made to the potential tumor antigens vFLIP or vCyclin in healthy donors or patients with disease.
How T-cell control is exercised over KSHV-associated malignancies is unclear, particularly in view of the immune evasion mechanisms used by the virus. Thus, LANA encodes an extensive acidic repeat sequence that inhibits efficient synthesis and proteasomal degradation of itself.13 This strategy, also used by the Epstein-Barr virus (EBV) homolog EBNA1,14 limits the supply of peptides for presentation to CD8+ T cells.15 KSHV-specific T-cell killing of infected endothelial cells has not been tested; however, such in vitro infected cells transiently express K516 that functions to down-regulate surface HLA class I and other costimulatory molecules. No studies have been performed on the ability of T cells to recognize KSHV-infected cells from MCD patients; however, PELs expressing reporter antigens are not recognized by cognate CD8+ T cells.17 Collectively, these studies question the ability of KSHV-specific CD8+ T cells to effectively control these virus-associated pathologies.
Because B cells express HLA class II, KSHV-specific CD4+ T cells could conceivably target PELs and KSHV-infected cells in MCD. In vitro studies of KSHV-infected tonsillar B cells have shown that activated CD4+ T cells can inhibit viral replication in these cells, albeit by an MHC-independent mechanism.18 Recently, it has been found that vIRF3 can inhibit promoter activity of the class II transactivator (CIITA), the master regulator of class II and other gene expression involved in the class II presentation pathway. Ectopic expression of vIRF3 decreased surface class II expression in B cells and small interfering RNA knockdown of vIRF3 in PELs increased class II expression.19 Whether KSHV-specific CD4 T cells can recognize PELs and the impact of vIRF3 interference with CIITA expression is unknown. Here, we examined the magnitude of the ex vivo T-cell response to KSHV latent antigens in a population of healthy carriers and derived LANA-specific CD4+ T-cell clones from these donors to probe recognition of HLA-matched PELs. The T cells were mostly unable to recognize these cells; however, bypassing the effects of vIRF3 through expression of CIITA restored T-cell recognition but not killing of the PELs.
Methods
Donors
Peripheral blood samples were collected from healthy adult Gambian donors negative for HIV1, HIV2, and hepatitis B and C infections. Plasma and PBMCs were separated and cryopreserved using standard procedures. Plasma was screened for KSHV-specific antibodies by testing against KSHV-infected PEL cell lines.20 Participants gave written informed consent in accordance with the Declaration of Helsinki; experiments were approved by The Gambia Government/MRC Laboratories Joint Ethics Committee and the South Birmingham Local Research Ethics Committee. HLA typing of donors was performed at the Anthony Nolan Trust by sequence-specific oligonucleotide PCR analysis.
ELISPOTs
ELISPOT assays were conducted on serial dilutions of PBMCs as described previously.21 PBMCs were stimulated using overlapping peptide libraries that spanned vFLIP, vCyclin, Kaposin, and LANA, excluding the acidic repeat. Peptides (Mimotopes) were synthesized based on the BC-1 sequence of KSHV (accession NC_003409) as 15mers overlapping by 10 and used in pools containing 10 to 13 peptides, each at a final concentration of 2 μg/mL. Statistical analysis was conducted using the Wilcoxon 2-sample test using SAS Version 8.2 software (SAS Institute).
T-cell cultures and clones
Antigen-specific cells from PBMC samples were stimulated with peptides at a concentration of 0.5 μg/mL for 7 days with IL-7 and IL-2,22 and reactive cells were selected on rechallenge with the peptides at a concentration of 0.5 μg/mL using an IFN-γ cell enrichment kit (Miltenyi Biotech). Enriched cells were cloned by limiting dilution, to establish a single cell–derived population with only 1 specificity, and they were maintained as described previously.23 Specificity of the clones was determined by incubating them with pools of peptides at a concentration of 0.5 μg/mL and then analyzing for IFN-γ release by ELISA (Endogen). Clones were then assayed with individual peptides within reactive pools to identify the peptide that induced function.
Construction of plasmids, transfection, and retroviral transduction of cells
LANA expression constructs were generated from BCBL-1 PEL DNA by PCR amplification of the regions flanking the LANA repeat sequence, namely, the first 1035 and final 735 nucleotides. These products were fused by PCR and cloned into pRTS1,24 adjacent to a bidirectional tetracycline-responsive promoter to create the LANAΔacid construct. A derivative of this construct included the first 240 nucleotides of the invariant chain gene fused to the 5′ terminus of the LANA sequence, creating the li LANAΔacid construct.
Retroviral constructs were engineered by cloning the cDNA encoding either CIITA or HLA-B*81 into the pLZRS retroviral vector, upstream from an internal ribosome entry site and the truncated nerve growth factor receptor (NGFR) gene. Virus was produced using the Phoenix packaging cell line, and PELs were transduced by incubating 400 000 cells with the virus and centrifuging at 3000g for 1 hour at 37°C. Transduced cells were magnetically sorted using NGFR-specific beads (Miltenyi Biotech). Surface HLA class II levels were measured on NGFR-expressing cells by costaining with anti-NGFR antibody (C40-1457, BD Biosciences) and an anti-class II MHC antibody (WR18, Serotec). Cells were analyzed on an Epics II flow cytometer (Beckman Coulter).
Protein preparations
LANA protein was enriched from BCBL-1 cells by extracting nuclear proteins from 500 × 106 cells,25 and these transferred into 0.2M NaCl and 0.05M Tris, pH 8.0, using PD-10 columns (GE Healthcare). Proteins were loaded onto a Mono Q anion exchange column (GE Healthcare), and fractions were eluted by fast protein liquid chromatography using a NaCl gradient from 0.2M to 1M. Eluted fractions containing LANA were identified by Western blot analysis. Nuclear extracts from the Burkitt lymphoma line DG-75 were similarly processed, and equivalent fractions were used as a control antigen.
T-cell recognition assays
Assays were conducted essentially as described previously.26 For each clone, 5000 T cells were incubated with 50 000 target cells in RPMI-10% FCS for 18 hours at 37°C in triplicate, and T-cell recognition was assessed by measuring IFN-γ secretion by ELISA. Target cells were either EBV-transformed B-lymphoblastoid cells (LCLs) made using the B95.8 virus, or PELs, both transfected with the above-mentioned plasmids as described previously,27 or transduced PELs. LANA expression from plasmids was induced by incubating LANAΔacid–transfected cells with 2 μg/mL doxycycline (dox), or li LANAΔacid–transfected cells with 3 ng/mL dox, for 72 hours. Antigen-sensitized targets were either incubated with peptides at a concentration of 5μM for 1 hour, or they were cultured for 18 hours with protein preparations in AIM-V medium (Invitrogen). Cells were extensively washed before use as targets.
CD4+ T-cell clones were tested for killing of target cells at known effector:target ratios in standard 5- and 12-hour chromium release cytotoxicity assays. Results are expressed as the percentage of specific lysis of the target line. Targets were LCLs or PELs pre-exposed for 1 hour to 5μM epitope-peptide or to an equivalent concentration of DMSO solvent as a control.
Western blot analysis
Cells were lysed in 9M urea and 0.075M Tris-HCl, pH 7.5, and then sonicated. Protein concentration was determined by Bradford assay (Bio-Rad Laboratories); 20 μg of protein was separated by SDS-PAGE and blotted onto nitrocellulose membranes using standard techniques. Blots were probed with antibodies specific to hemagglutinin (clone 3F10, Roche Diagnostics); LANA (clone AT4C11, Abnova); vIRF3 (CM-A807, Abcam); CIITA, HLA-DR, and calregulin (Santa Cruz Biotechnology); and actin (AC-74, Sigma-Aldrich); and these were detected using an appropriate anti-species HRP-conjugated antibody, followed by ECL detection (GE Healthcare).
Results
Characterization of healthy donor T-cell responses to KSHV latent antigens
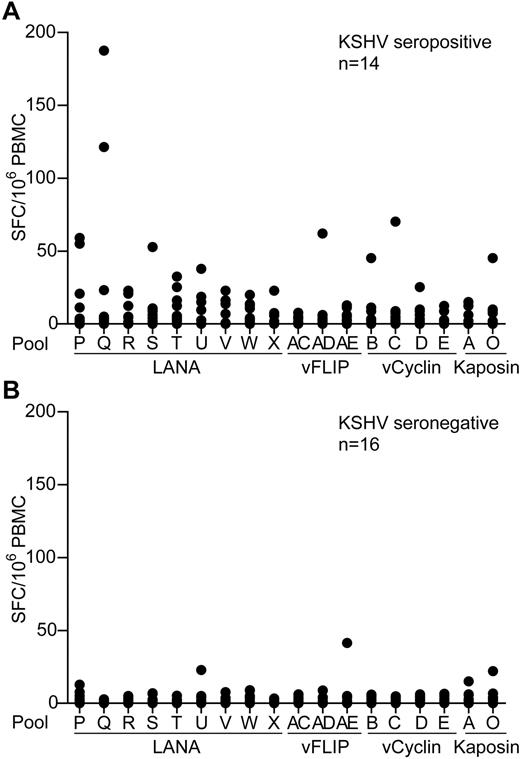
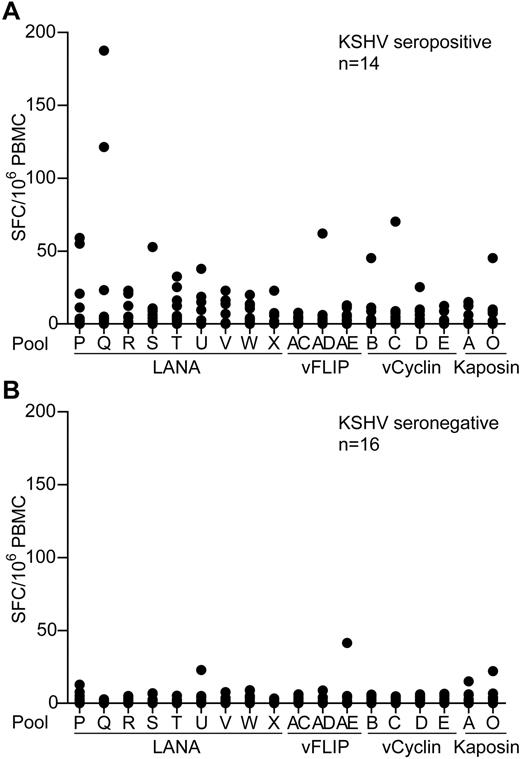
To identify and quantify KSHV latent antigen-specific T-cell responses, PBMC samples from healthy donors living in a region where KSHV infection has a relatively high incidence were screened by IFN-γ ELISPOT for reactivity to these antigens. PBMCs were assayed against pools of overlapping peptides spanning these proteins that included 9 LANA peptide pools, excluding the repeat sequences, 3 vFLIP, 4 vCyclin, and 2 Kaposin pools. Figure 1A-B shows summaries of the ELISPOT responses to the individual peptide pools made by 14 seropositive and 16 seronegative donors. Most donors' PBMCs showed no or very weak responses to stimulation with the peptides, irrespective of their KSHV serology status. However, several seropositive donors made low responses to the LANA peptide pools, with weak, infrequent responses to the vFLIP, vCyclin, and Kaposin peptides. Statistical differences were seen when comparing responses made by seropositive versus seronegative donors to pools Q, T, and W (P ≤ .03); however, the weakness of these responses questions the biologic significance of these statistics.
KSHV latent antigen-specificELISPOTanalysis of PBMCs from KSHV seropositive and seronegative donors. PBMCs from 14 KSHV seropositive donors (A) and 16 KSHV seronegative donors (B) were tested by IFN-γ ELISPOT against pools of overlapping peptides that spanned the KSHV latent antigens. Results are reported as number of spot-forming cells (SFC) per million PBMCs. Background responses to the DMSO control have been subtracted from the values presented; mean values were 10 and 3 SFC/million PBMCs for the seropositive and seronegative donors, respectively.
KSHV latent antigen-specificELISPOTanalysis of PBMCs from KSHV seropositive and seronegative donors. PBMCs from 14 KSHV seropositive donors (A) and 16 KSHV seronegative donors (B) were tested by IFN-γ ELISPOT against pools of overlapping peptides that spanned the KSHV latent antigens. Results are reported as number of spot-forming cells (SFC) per million PBMCs. Background responses to the DMSO control have been subtracted from the values presented; mean values were 10 and 3 SFC/million PBMCs for the seropositive and seronegative donors, respectively.
Identification of KSHV-specific T-cell responses
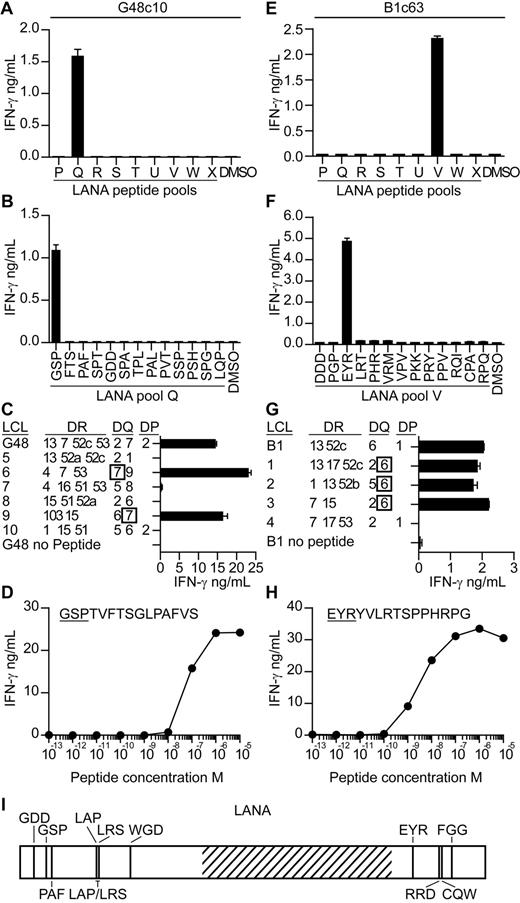
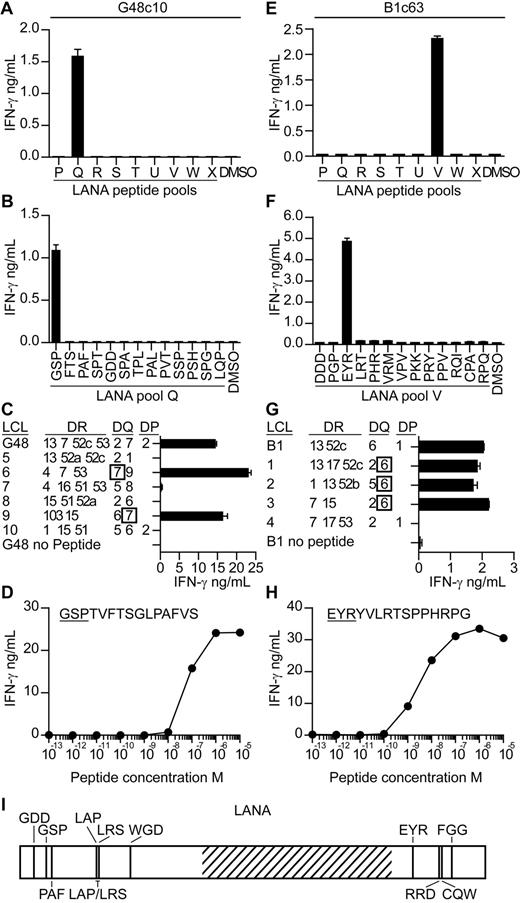
T-cell clones were subsequently generated from seropositive donors' PBMCs to map KSHV epitopes and to investigate their ability to recognize antigen-expressing cells. We focused on identifying epitopes within LANA, because the ELISPOT analysis suggested LANA induced the most frequent, albeit weak, responses. PBMCs were stimulated with LANA peptides for 7 days,22 after which LANA-specific T cells were sorted, and single cell–derived clones were generated. The specificity of the clones was identified by incubating them with the LANA peptide pools and then assaying for IFN-γ secretion. Figure 2 shows mapping studies from 2 representative clones. Clone c10, derived from donor G48, released IFN-γ when stimulated with pool Q peptides, and within this pool, peptide GSPTVFTSGLPAFVS induced this response (Figure 2A-B). Clones are subsequently identified by the first 3 letters of their cognate epitope-peptide. Clones were tested for expression of CD4 or CD8 by flow cytometry, and all but one specificity expressed CD4 (data not shown). We concentrated on studying the CD4 clones as these gave a greater range of specificities with which to probe recognition of antigen-expressing cells. The HLA restriction of the clones was identified by testing them against peptide-sensitized partially HLA class II–matched EBV transformed B-cell lines (LCLs) and then identifying which LCLs induced IFN-γ secretion. As shown in Figure 2C, peptide-sensitized HLA DQ7-matched LCLs stimulated GSP-specific function, indicating this molecule presented the peptide. The functional avidity of this clone was assessed by testing it against LCL sensitized with 10-fold dilutions of GSP peptide and measuring the concentration which elicited 50% maximal interferon release. As shown in Figure 2D, the GSP-specific clone had a functional avidity in the order of 1 × 10−7M.
Characterization of LANA-specific CD4+ T-cell clones. T-cell clones from donor G48 and B1 were screened for IFN-γ secretion by ELISA when incubated with the pools of LANA peptides (A,E) and then the individual peptides within the reactive pools (B,F). The HLA restriction of the clones was then determined by testing the clones for IFN-γ release when challenged with partially HLA-matched LCLs sensitized with the clone's cognate peptide (C,G). Functional avidity of the clones was determined by incubating the clones with LCLs sensitized with 10-fold dilutions of cognate peptide and then assaying for IFN-γ secretion (D,H). Map of novel epitopes within LANA. Hatched region represents the acidic repeat region of LANA (I).
Characterization of LANA-specific CD4+ T-cell clones. T-cell clones from donor G48 and B1 were screened for IFN-γ secretion by ELISA when incubated with the pools of LANA peptides (A,E) and then the individual peptides within the reactive pools (B,F). The HLA restriction of the clones was then determined by testing the clones for IFN-γ release when challenged with partially HLA-matched LCLs sensitized with the clone's cognate peptide (C,G). Functional avidity of the clones was determined by incubating the clones with LCLs sensitized with 10-fold dilutions of cognate peptide and then assaying for IFN-γ secretion (D,H). Map of novel epitopes within LANA. Hatched region represents the acidic repeat region of LANA (I).
For the second representative clone, c63 from donor B1 (Figure 2E-H), its epitope-peptide was found within peptide pool V to be the peptide EYRYVLRTSPPHRPG. This peptide was presented by HLA DQ6-expressing LCLs, and the clone had a functional avidity of 1 × 10−8M. Overall, 51 peptide-specific clones were established from 4 donors, specific to 1 of 13 epitopes (Table 1; Figure 2I). In addition, we checked the cytotoxic potential of the clones for which we identified HLA restrictions, assaying against peptide-sensitized LCLs in standard chromium release cytotoxicity assays. With the exception of clones G48 PAF and G29 PAF, all were cytotoxic (data not shown).
Recognition of LANA-expressing targets
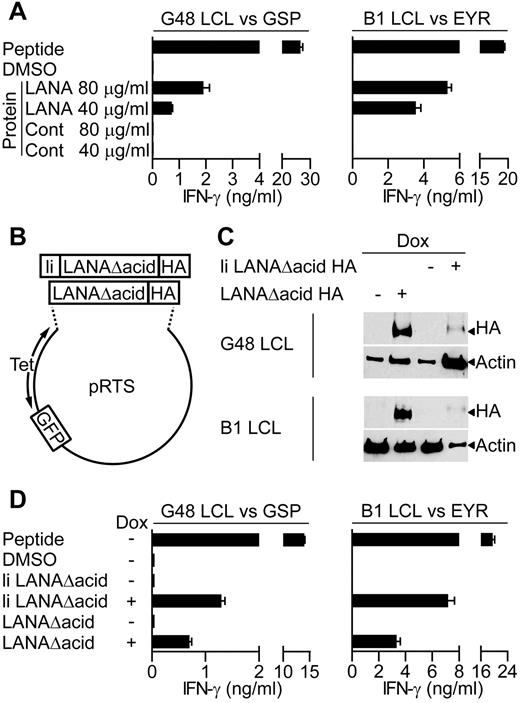
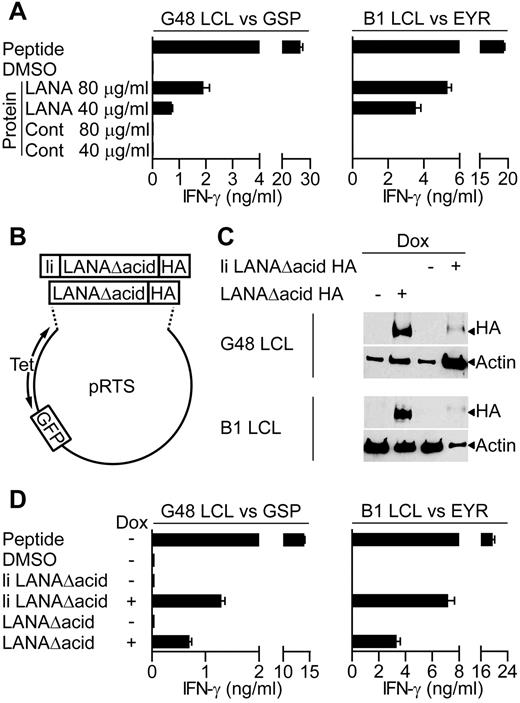
Because a major cellular target for KSHV infection are B lymphocytes, we determined whether the LANA-specific T-cell clones recognized antigen-expressing B lymphocytes. Because in vitro infection of primary B cells with KSHV is difficult, 2 independent assays were used to assess recognition of B cells exposed to or expressing LANA antigen. Here, LCLs were used as targets, because they are competent for processing and presentation of class II epitopes. Initially, we asked whether the clones recognized donor B1 or G48 LCLs that had been incubated for 18 hours with enriched LANA protein extracted from BCBL-1 cells, or a control protein preparation. For recognition of the LCL to occur, these targets must take up the LANA protein and process and present the epitope-peptide from it to the T-cell clones. Figure 3A shows representative examples of such assays using GSP- and EYR-specific clones. In both cases, the clones recognized the LCLs incubated with enriched LANA protein but not those incubated with the control protein. This pattern was seen for all of the clones tested.
LANA-specific CD4+ T-cell recognition of LCLs expressing LANA antigens. (A) IFN-γ production by GSP- or EYR-specific T cells incubated with G48 LCLs or B1 LCLs, respectively, fed either an enriched LANA or control (Cont) protein preparation. (B) Diagram of recombinant plasmids that expressed the LANA constructs and GFP reporter under the control of the tetracycline-responsive promoter. (C) Western blot analysis of recombinant LANA expressed from transfected G48 or B1 LCLs treated with dox. Blots were probed with a hemagglutinin (HA)–specific antibody to detect recombinant LANA. (D) IFN-γ secretion from GSP- or EYR-specific T cells incubated with G48 or B1 LCLs, respectively, either sensitized with 5μM cognate peptide or induced to express recombinant LANA by dox treatment.
LANA-specific CD4+ T-cell recognition of LCLs expressing LANA antigens. (A) IFN-γ production by GSP- or EYR-specific T cells incubated with G48 LCLs or B1 LCLs, respectively, fed either an enriched LANA or control (Cont) protein preparation. (B) Diagram of recombinant plasmids that expressed the LANA constructs and GFP reporter under the control of the tetracycline-responsive promoter. (C) Western blot analysis of recombinant LANA expressed from transfected G48 or B1 LCLs treated with dox. Blots were probed with a hemagglutinin (HA)–specific antibody to detect recombinant LANA. (D) IFN-γ secretion from GSP- or EYR-specific T cells incubated with G48 or B1 LCLs, respectively, either sensitized with 5μM cognate peptide or induced to express recombinant LANA by dox treatment.
In the second series of recognition experiments, LANA expression plasmids (Figure 3B) were transfected into B1 and G48 LCLs, and these LCLs were used as targets. Two LANA constructs were generated, both with the acidic repeat domain removed (denoted as LANAΔacid) because this increases levels of protein translation,13 and both included the influenza HA antibody-epitope at the carboxy terminus to allow detection. In addition, 1 construct had the first 80 amino acids of the HLA class II invariant chain fused to it (denoted as li LANAΔacid) to redirect protein to the endosomal compartment, allowing direct loading of peptides onto HLA class II molecules.28 LANA construct expression was controlled from a bidirectional tetracycline regulated promoter, allowing inducible expression of LANA and GFP as a reporter.
Expression of the LANA constructs in the LCLs was induced with the tetracycline analog dox for 72 hours, and then expression examined by Western blot analysis. Figure 3C shows induced expression of both constructs, although the expression of li LANAΔacid seems weak, this is probably a consequence of lower proportions of cells being transfected with this construct as determined by green fluorescent protein (GFP) expression. Thus, 3.6% of G48 LCLs and 2.6% of B1 LCLs contained the li LANAΔacid plasmid compared with 44.6% and 50% for the LANAΔacid construct, respectively. G48-transfected cells were used as targets in recognition assays with the HLA DQ7-restricted GSP-specific clone and for assaying IFN-γ production. Nontransfected LCLs sensitized with GSP-peptide were used as a positive control, and LCLs sensitized with the peptide solvent DMSO or uninduced LCLs served as negative controls. In representative results from 3 experiments, peptide-sensitized LCLs induced high levels of IFN-γ secretion from the clones, with little recognition of the DMSO or uninduced LCLs (Figure 3D left). li LANAΔacid-expressing cells were well recognized, probably because of the endosomal routing of this protein. Importantly, LCLs expressing LANAΔacid stimulated IFN-γ release from the GSP-specific clone.
Recognition assays using B1 LCLs transfected with the LANAΔacid constructs gave similar results. Figure 3D (right) shows representative results of assays using 1 B1 clone of 5, tested on 4 occasions. The HLA DQ6-restricted EYR-specific clone showed good recognition of the li LANAΔacid-expressing LCLs and the LANAΔacid-expressing LCLs. All clones tested from B1 were found to be capable of recognizing LCLs expressing LANAΔacid (data not shown). In summary, all LANA-specific clones tested were found to be capable of recognizing LCLs expressing LANAΔacid.
Recognition of PEL cell lines by LANA-specific CD4+ T cells
We next determined whether the LANA-specific CD4+ T-cell clones were capable of recognizing KSHV-associated malignancies, in this case PEL cell lines. These malignancies are of B-cell origin, express variable levels of surface HLA class II29 and express KSHV latent genes, including LANA. The PELs used in this study and their HLA class II types are shown in Table 2.
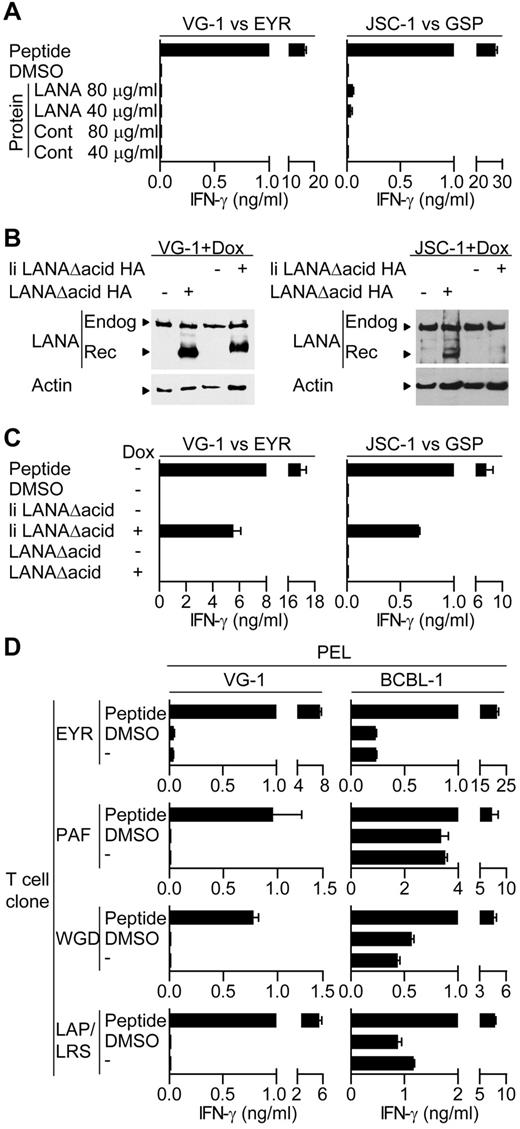
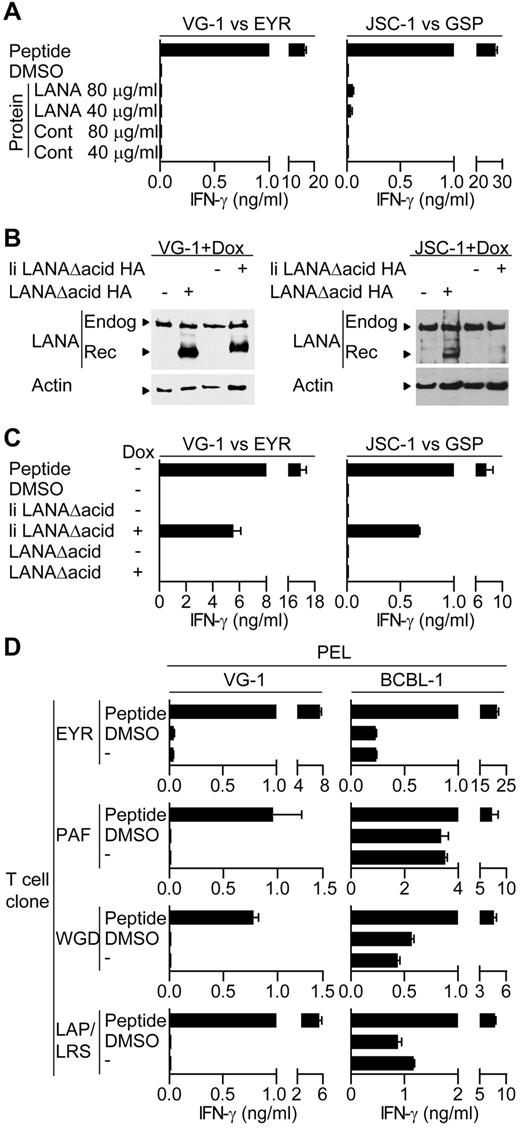
Initially, we tested the ability of HLA-matched T-cell clones to recognize PEL cell lines incubated with either LANA or the control protein preparation. Figure 4A shows representative results using EYR-specific and GSP-specific clones assayed against VG-1– and JSC-1–sensitized PELs, respectively. Peptide-sensitized targets induced good levels of IFN-γ secretion by the clones; however, little or no recognition of the PELs sensitized with DMSO or the control antigen was seen, indicating no recognition of LANA expressed from the endogenous KSHV genome. Furthermore, in contrast to the LCL experiments, PELs sensitized with LANA protein induced no obvious additional recognition by the clones, suggesting the PELs were unable to process and present exogenous LANA protein. This pattern of reactivity was seen using all clones tested against the VG-1, BC-1, and JSC-1 PEL lines.
LANA-specific CD4+ T cell recognition of PELs. (A) IFN-γ production by EYR- or GSP-specific T cells incubated with VG-1 or JSC-1 PELs, respectively, fed either an enriched LANA protein or a control protein preparation. (B) Western blot analysis of endogenous and recombinant LANA expression from transfected VG-1 or JSC-1 PELs treated with dox. Blots were probed with a LANA-specific antibody. (C) IFN-γ secretion from EYR- or GSP-specific T cells incubated with VG-1 or JSC-1 PELs, respectively, either sensitized with 5μM cognate peptide or induced to express recombinant LANA by dox treatment. (D) HLA DQ6-expressing PELs VG-1 (left) or BCBL-1 (right) were incubated with HLA DQ6-restricted T cells specific for EYR, PAF, WGD, or LAP/LRS, and then IFN-γ release was quantified.
LANA-specific CD4+ T cell recognition of PELs. (A) IFN-γ production by EYR- or GSP-specific T cells incubated with VG-1 or JSC-1 PELs, respectively, fed either an enriched LANA protein or a control protein preparation. (B) Western blot analysis of endogenous and recombinant LANA expression from transfected VG-1 or JSC-1 PELs treated with dox. Blots were probed with a LANA-specific antibody. (C) IFN-γ secretion from EYR- or GSP-specific T cells incubated with VG-1 or JSC-1 PELs, respectively, either sensitized with 5μM cognate peptide or induced to express recombinant LANA by dox treatment. (D) HLA DQ6-expressing PELs VG-1 (left) or BCBL-1 (right) were incubated with HLA DQ6-restricted T cells specific for EYR, PAF, WGD, or LAP/LRS, and then IFN-γ release was quantified.
Because we were unable to demonstrate T-cell recognition of the PELs, we overexpressed LANA in these cells to force recognition. Here, PELs were transfected with the inducible LANAΔacid expression plasmids used above, and expression was induced with dox. Figure 4B shows representative Western blots of lysates from the transfected VG-1 or JSC-1 cells probed with a LANA-specific antibody, detecting both the endogenous and induced LANA proteins after dox induction. Aliquots of these cells and uninduced cells sensitized with the cognate peptide were then used as targets in recognition assays. Figure 4C shows representative results of at least 2 independent assays using the HLA DQ6-restricted EYR-specific clone and the HLA DQ7-restricted GSP-specific clone, assayed against VG-1 and JSC-1, respectively. Again, clones recognized the peptide-sensitized PELs; however, no or very weak recognition of either DMSO or uninduced PELs was observed. PELs expressing the endosomally directed li LANAΔacid construct induced responses; however, in contrast to the LCL experiments, PELs expressing LANAΔacid induced no additional T-cell recognition. Other B1 clones that were HLA-matched with VG-1– and the GSP-specific clone when tested against a second HLA DQ7-matched PEL BC-1 showed the same pattern of reactivity. No recognition of the endogenous or overexpressed LANAΔacid was observed; only cells sensitized with cognate peptide or expressing li LANAΔacid stimulated responses from the T cells (data not shown).
In contrast to the above-mentioned pattern of PEL cell recognition, the BCBL-1 line consistently induced IFN-γ secretion when incubated with HLA-matched T cells. Figure 4D shows examples of duplicate assays using HLA DQ6-restricted T-cell clones derived from B1 assayed in parallel against BCBL-1 and VG-1, cells that also express HLA DQ6. In all cases, unmanipulated VG-1 cells induced little if any IFN-γ secretion, whereas BCBL-1 cells always induced IFN-γ secretion.
Expression of CIITA increases HLA class II expression on PELs and restores recognition by LANA-specific CD4+ T cells
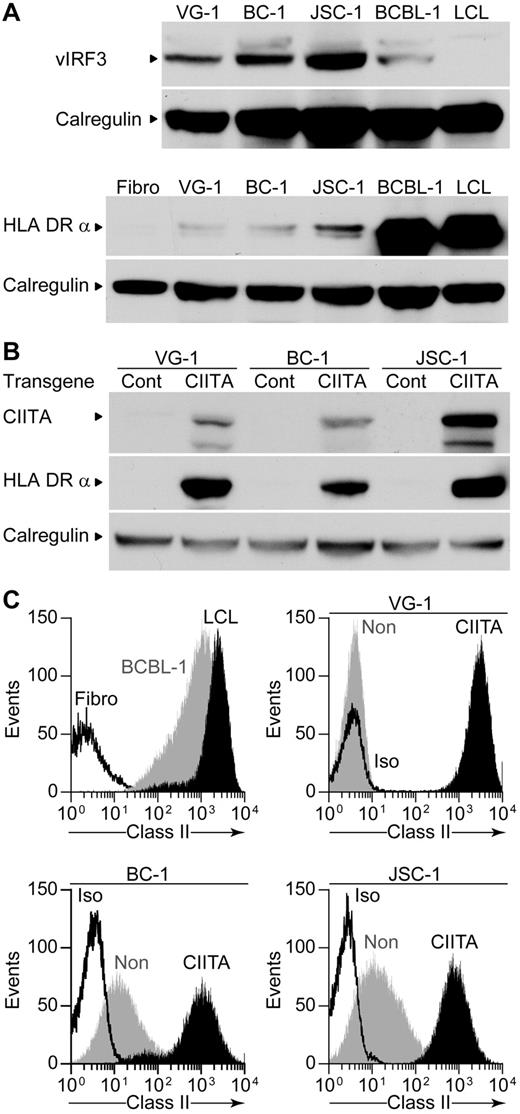
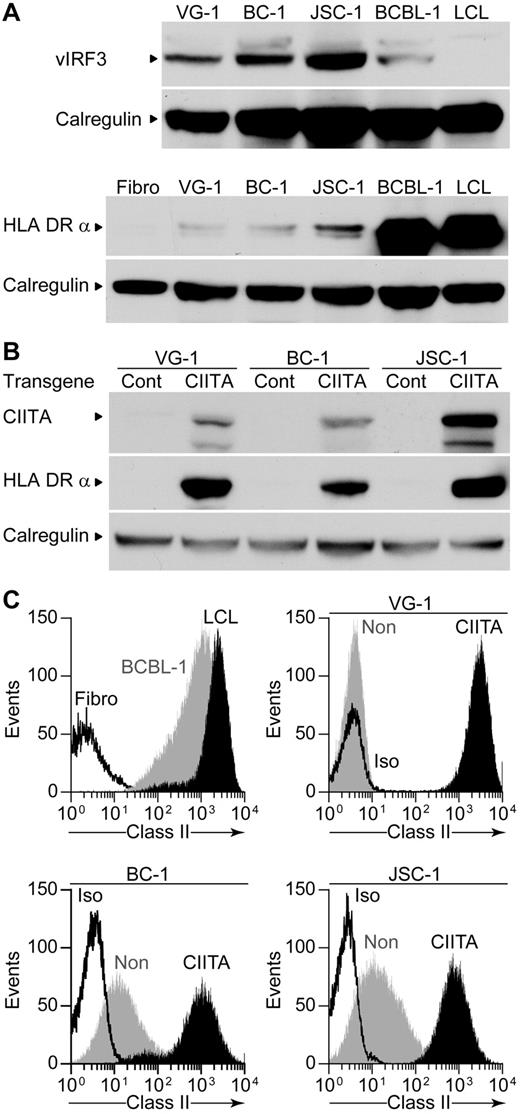
The KSHV-encoded vIRF3 protein, which is expressed in PELs, has been found to inhibit the B cell–specific PIII promoter and IFN-γ–responsive PIV promoter of the CIITA,19 which drives expression of HLA class II and other genes. Western blot analysis of vIRF3 expression in the different PEL lines used showed all PELs expressed vIRF3; interestingly, BCBL-1 showed the lowest level of expression of this protein (Figure 5A). We also probed the blots for HLA-DR and found that BCBL-1 cells expressed high amounts, whereas minimal levels were observed in the other PELs, potentially providing an explanation for the poor recognition of these latter PELs by the LANA-specific CD4+ T cells. We were unable to up-regulate surface class II expression on the PELs by treating with IFN-γ (data not shown), probably a consequence of the inhibition of the PIV promoter by vIRF3.
vIRF3 and HLA class II expression by PELs and expression of CIITA and HLA class II after CIITA transduction. (A) Western blot analysis of lysates from PELs probing for vIRF3 expression (top) or for the HLA DRα chain expression (bottom) and calregulin expression as a loading control. (B) Western blot analysis of lysates from Cont–transduced or CIITA-transduced PELs, probing for CIITA, HLA-DRα, and calregulin as a loading control. (C) Flow cytometry analysis of surface HLA class II levels on BCBL-1, an LCL or a fibroblast (top left), VG-1 transductants (top right), BC-1 transductants (bottom left), and JSC-1 transductants (bottom right). CIITA-transduced PELs are shown in black, nontransduced PELs in gray, and isotype control staining is open.
vIRF3 and HLA class II expression by PELs and expression of CIITA and HLA class II after CIITA transduction. (A) Western blot analysis of lysates from PELs probing for vIRF3 expression (top) or for the HLA DRα chain expression (bottom) and calregulin expression as a loading control. (B) Western blot analysis of lysates from Cont–transduced or CIITA-transduced PELs, probing for CIITA, HLA-DRα, and calregulin as a loading control. (C) Flow cytometry analysis of surface HLA class II levels on BCBL-1, an LCL or a fibroblast (top left), VG-1 transductants (top right), BC-1 transductants (bottom left), and JSC-1 transductants (bottom right). CIITA-transduced PELs are shown in black, nontransduced PELs in gray, and isotype control staining is open.
We attempted to inhibit vIRF3 function in the PELs using previously described small interfering RNAs and protocols,19,30 but we were unable to show any reduction of vIRF3 protein (data not shown). Instead, because vIRF3 interferes with the CIITA promoters, we transduced the PELs with a retrovirus expressing either CIITA or a control construct from a different promoter. Lysates of the CIITA-transduced cells analyzed by Western blot showed expression of CIITA and proteins that it transactivates, namely, CD74 (data not shown) and HLA-DR (Figure 5B). Expression of surface levels of class II was measured by flow cytometry on the transduced PELs and compared with levels on BCBL-1, an LCL, and class II negative fibroblasts (Figure 5C). Control-transduced cells showed little difference in surface class II expression compared with nontransduced cells (data not shown); however, all CIITA-transductants expressed increased levels of class II, comparable to those expressed by the LCL or BCBL-1.
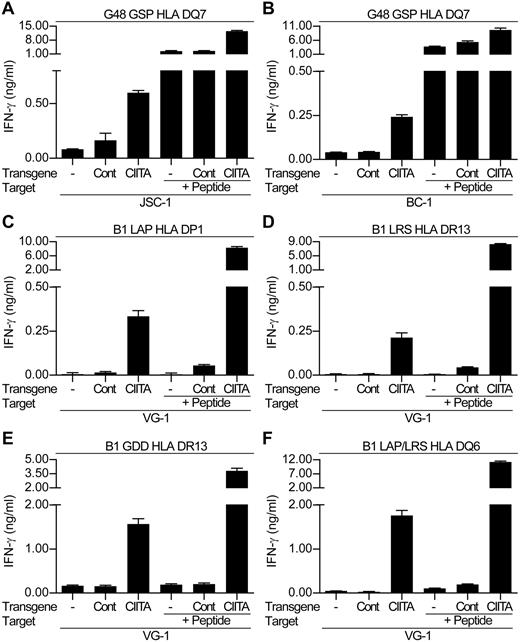
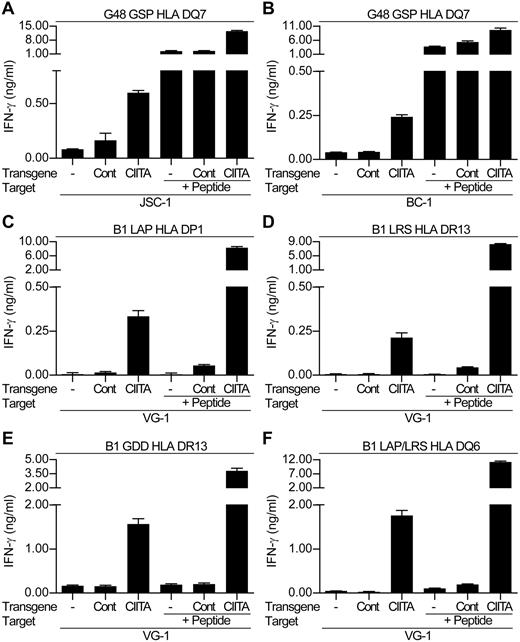
Because the CIITA-transduced PELs showed increased levels of surface class II, we re-examined the ability of the LANA-specific CD4+ T-cell clones to recognize these cells. Figure 6 shows representative results from 1 of 3 experiments comparing T-cell recognition of the nontransduced and control-transduced PELs versus the CIITA-transduced PELs. Assaying the DQ7-restricted GSP-specific clone against nontransduced and control-transduced JSC-1 and BC-1 cells induced little or no IFN-γ release (Figure 6A and B, respectively). Similarly, HLA-mismatched LANA-specific clones showed little recognition of the transduced PELs (data not shown). However, when the CIITA-transduced PELs were used as targets with the HLA-matched GSP-specific clone, the clones secreted IFN-γ, indicating they could now recognize these PELs.
LANA-specific CD4+ T-cell recognition of CIITA or control-transduced PELs. The HLA DQ7-restricted GSP-specific clone was incubated in recognition assays with either nontransduced (-), Cont-transduced, or CIITA-transduced (A) JSC-1 or (B) BC-1 PELs. VG-1–nontransduced, control-transduced, or CIITA-transduced PELs were incubated with either HLA DP1-restricted LAP-specific clones (C), HLA DR13-restricted LRS-specific clones (D), HLA DR13-restricted GDD-specific clones (E), or HLA DQ6-restricted LAP/LRS-specific clones (F). In each case, aliquots of target cell lines were also sensitized with 0.1μM of the cognate peptide as controls. Note that this is a lower concentration than that used in Figure 4, which in some cases was insufficient to sensitize targets.
LANA-specific CD4+ T-cell recognition of CIITA or control-transduced PELs. The HLA DQ7-restricted GSP-specific clone was incubated in recognition assays with either nontransduced (-), Cont-transduced, or CIITA-transduced (A) JSC-1 or (B) BC-1 PELs. VG-1–nontransduced, control-transduced, or CIITA-transduced PELs were incubated with either HLA DP1-restricted LAP-specific clones (C), HLA DR13-restricted LRS-specific clones (D), HLA DR13-restricted GDD-specific clones (E), or HLA DQ6-restricted LAP/LRS-specific clones (F). In each case, aliquots of target cell lines were also sensitized with 0.1μM of the cognate peptide as controls. Note that this is a lower concentration than that used in Figure 4, which in some cases was insufficient to sensitize targets.
Parallel experiments were conducted against the VG-1–transduced cells using a representative panel of clones from donor B1, restricted through HLA DP1, DR13, and DQ6 (Figure 6C-F). The clones showed little recognition of the nontransduced and control-transduced VG-1 cells, whereas an HLA-mismatched clone showed minimal recognition of the VG-1 transductants (data not shown). However, all clones were capable of recognizing the CIITA-transduced VG-1 PEL. Thus, the LAP- and LRS-specific clones showed intermediate recognition of the CIITA-transduced PELs, whereas the GDD-specific and LAP/LRS-specific clones showed strong recognition of this target.
We also checked whether the clones showed cytotoxic function when challenged with the PELs in cytotoxicity assays. Here, we used the CIITA-transduced PEL VG-1 and the unmanipulated BCBL-1 for which we had HLA-matched cytotoxic effectors. Although the clones showed good cytotoxicity against peptide-sensitized LCLs, in parallel assays they showed weaker, if any, killing against the peptide-sensitized PELs and no killing of CIITA-transduced VG-1 or unmanipulated BCBL-1 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), despite the clones secreting IFN-γ when challenged with these latter 2 targets.
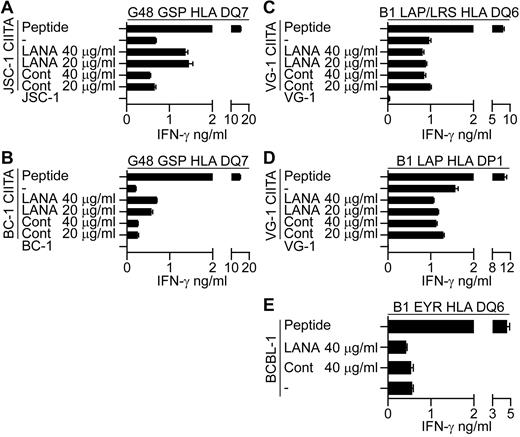
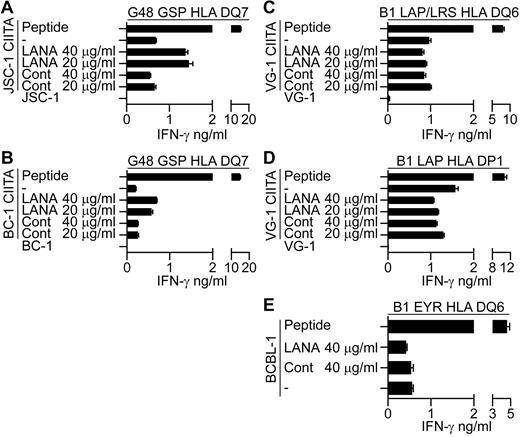
Preliminary experiments were conducted to identify the route of presentation of LANA epitopes in the CIITA-transduced PELs. Here, evidence was sought as to whether the epitopes are directly processed from endogenous LANA or whether exogenously acquired LANA can be taken up and processed and presented to the T cells. CIITA-transduced PELs were fed enriched LANA or control protein preparations for 18 hours and used as targets in recognition assays. As controls, LCLs were fed with the proteins in parallel assays and LANA fed LCLs were recognized by the T cells in each case (data not shown). CIITA-transduced JSC-1 and BC-1 PELs fed LANA protein induced increased recognition by the G48 GSP-specific T cells (Figure 7A-B, 1 representative experiment of 3), suggesting that recognition of these CIITA-transduced PELs may be due at least in part to reprocessing of exogenously acquired LANA protein. However, when LANA fed CIITA-transduced VG-1 PELs were assayed against B1 LAP/LRS or B1 LAP effectors, no increase in recognition was observed (Figure 7C-D, 1 representative experiment of 3). Similarly, unmanipulated BCBL-1 targets that induce IFN-γ secretion from the B1 DQ6 restricted clone showed no increase in recognition when fed LANA protein (Figure 7E, 1 representative clone of 4). These latter 2 results suggest that these PELs do not present epitopes from exogenous LANA, implying a role for direct presentation of endogenous LANA epitopes in these cases.
LANA-specific CD4+ T-cell recognition of CIITA-transduced JSC-1, BC-1, and VG-1 PELs or unmanipulated BCBL-1 fed enriched LANA-protein. CIITA-transduced JSC-1, BC-1, or VG-1 PELs or the unmanipulated BCBL-1 PELs were fed enriched LANA protein or a control protein preparation for 18 hours before being washed and incubated with the appropriate HLA-matched T-cell clone, and then IFN-γ secretion was measured. (A-B) Protein-fed JSC-1– and BC-1–transduced PELs, respectively, assayed against the HLA DQ7-restricted GSP-specific clone. Protein-fed VG-1–transduced PELs assayed against HLA DQ6-restricted LAP/LRS-specific clones (C) or HLA DP1-restricted LAP-specific clones (D). (E) Protein-fed BCBL-1 cells assayed against the HLA DQ6-restricted EYR-specific clone.
LANA-specific CD4+ T-cell recognition of CIITA-transduced JSC-1, BC-1, and VG-1 PELs or unmanipulated BCBL-1 fed enriched LANA-protein. CIITA-transduced JSC-1, BC-1, or VG-1 PELs or the unmanipulated BCBL-1 PELs were fed enriched LANA protein or a control protein preparation for 18 hours before being washed and incubated with the appropriate HLA-matched T-cell clone, and then IFN-γ secretion was measured. (A-B) Protein-fed JSC-1– and BC-1–transduced PELs, respectively, assayed against the HLA DQ7-restricted GSP-specific clone. Protein-fed VG-1–transduced PELs assayed against HLA DQ6-restricted LAP/LRS-specific clones (C) or HLA DP1-restricted LAP-specific clones (D). (E) Protein-fed BCBL-1 cells assayed against the HLA DQ6-restricted EYR-specific clone.
Discussion
Studies of the immunosuppressed have indicated that T-cell immunity is crucial to maintenance of an appropriate virus host balance in KSHV infection,3,31 yet few have examined the size and repertoire of responses to the well-defined KSHV latent antigens in healthy carriers who are controlling their infection. We examined the T-cell response in such donors to the KSHV latent proteins LANA, vFLIP, vCyclin, and Kaposin by assessing ELISPOT responses to libraries of overlapping peptides. This gives a global estimation of the T-cell response to these antigens across all HLA types, avoiding any bias by selecting donors with particular HLA types. These ex vivo responses were weak and substantially lower than those made to the other human γ-herpesvirus, EBV, by these and other donors from this population (data not shown; ref. 32). However, KSHV does not encode homologs of the immunodominant latent EBV EBNA3 family of proteins, but it does encode LANA, a functional homolog of EBNA1. Responses to LANA seem considerably weaker than those made to EBNA1 in donors from this32 and other populations.33,34 These KSHV-specific responses were also lower than those reported in similar studies using individuals co-infected with HIV on HAART therapy, yet they showed a similar immunodominance profile with epitopes from LANA more frequently recognized than Kaposin.9,10 These responses did seem more frequent than those described in HIV-infected donors with KS before HAART2 or in transplant patients with KS before changing their immunosuppressive regime.35
Because the ex vivo T-cell responses were weak, we generated T-cell clones for further study. Here, PBMCs were briefly expanded by stimulating with the LANA library of 15mer peptides before generating single-cell clones that established predominantly CD4-expressing clones. We cannot exclude that the culture conditions preferentially expanded CD4 over CD8 specificities, indeed others have found 15mer peptides preferentially stimulate CD8 cells over CD4.36 However, our findings show some contrast to what may be expected based on studies of ex vivo stimulation of donors' PBMCs with overlapping LANA peptides that resulted in stimulation of mostly CD8+ T cells.9 Furthermore, ex vivo stimulation of PBMCs with dendritic cells sensitized with overlapping LANA peptides has been shown to stimulate CD8+ T-cell responses.5
Using these LANA-specific CD4+ clones to probe recognition of LANA-expressing cells, we found that EBV-transformed B cells, cells that have an intact class II antigen processing pathway, could efficiently process and present LANA antigens. However, when the clones were tested against most PELs, cells that natively express LANA, no recognition was seen, nor when the protein was overexpressed or fed to the PELs. The inhibition of CIITA transcription by vIRF3 provides an explanation for the poor recognition of these cells by the CD4+ T cells. CIITA binds to 4 transacting factors that act as an enhanceosome, bound to regulatory modules within the promoter regions of genes associated with the class II pathway such as HLA-DP, HLA-DQ, HLA-DR, the invariant chain and the nonclassical class II molecules HLA-DM and HLA-DO.37 When bound to the enhanceosome, CIITA promotes transcription by recruiting and activating transcription machinery. Inhibiting CIITA expression represents an effective strategy to restrict expression of many components of the HLA class II processing pathway, and its importance is highlighted by the observations that the B-cell tropic pathogen EBV and others target expression of this gene.38,39
Interestingly, BCBL-1 cells relative to the other PELs expressed lower amounts of vIRF3, had higher levels of surface class II expression and reliably induced IFN-γ secretion from HLA-matched LANA-specific T cells. How representative these different PEL lines are of in vivo malignancies or in vivo–infected B cells, for that matter, is unclear. Indeed, KSHV protein expression in cultured cells has been suggested to not accurately reflect the in vivo situation.40 In vivo, all PEL cells have been shown to express vIRF341 ; however, the level of expression and impact on class II levels on these PEL cells will be of interest to determine. Nevertheless, our T-cell recognition studies are consistent with the idea that vIRF3 inhibition of CIITA promoter usage is a factor in preventing efficient class II processing and presentation of LANA epitopes. The observation that PELs such as VG-1 express similar levels of vIRF3 compared with BC-1 or JSC-1 but have lower levels of surface class II may suggest other factors are involved in subverting class II processing in PELs. In this context, vFLIP has been proposed to be an inhibitor of autophagy,42 known to be important in the generation of CD4 epitopes,43 and it may then restrict presentation of KSHV-epitopes. Nevertheless, bypassing the CIITA promoter blockage by vIRF3 through the ectopic expression of CIITA restored expression of class II and other CIITA targets in the PELs sufficient to overcome any other potential inhibitory mechanisms.
When the CIITA-transduced PEL VG-1 or BCBL-1 PELs were used as targets in cytotoxicity assays, they were not lysed by their cognate LANA-specific CD4+ T-cell clones, despite the clones recognizing these targets and producing IFN-γ. KSHV is known to encode several antiapoptotic mechanisms, most notably vFLIP, that inhibits Fas-mediated apoptosis by preventing caspase activation.44 It is currently unclear whether T cell–directed Fas-mediated effector mechanisms can kill PELs. Indeed, other cytotoxic pathways may be more effective, such as those induced by granzymes, particularly granzyme A, that are less dependent on caspase activation.45 Alternatively, coadministration of agents such as azidothymidine that sensitizes PELs in vitro to apoptosis mediated by tumor necrosis factor–related apoptosis-inducing ligand,46 a ligand that can be expressed by T cells, may be worth exploring. Although the T-cell clones did not kill these PELs, IFN-γ has been shown to inhibit virus production from PELs in vitro.47
vIRF3 is a multifunctional protein being able to inhibit type I interferon signaling,30 is required for the survival of PELs,48 inhibits p53 function,41 and inhibits CIITA expression.19 This multifunctionality makes vIRF3 an attractive therapeutic target not only in PEL but potentially other KSHV-associated diseases. Thus, vIRF3 is expressed in KSHV-infected B cells in MCD41 ; targeting this gene's function would allow the development of immunotherapeutic treatments for this aggressive disease. This is of particular relevance because the incidence of MCD is increasing in HIV-infected patients on HAART, and immune competence seems to be relatively conserved in these donors.11,49
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the field workers of the MRC laboratories in The Gambia for help in recruiting donors.
A.D.H. and this work are funded by a New Investigator Award from the Medical Research Council United Kingdom (G0501074).
Authorship
Contribution: S.S., Y.J.J., J.Z., M.M.A., and A.D.H. performed experiments; T.d.S., K.L.F., and S.R.-J. implemented recruitment and performed clinical assessments of donors; C.B. provided essential reagents; A.D.H. designed the experiments; and S.S. and A.D.H. interpreted the results and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.M.A. is Incepta Vaccine Ltd, Zirabo, Savar, Dhaka, Bangladesh.
Correspondence: Andrew D. Hislop, School of Cancer Sciences and MRC Centre for Immune Regulation, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom; e-mail: a.d.hislop@bham.ac.uk.