Abstract
The Fanconi anemia (FA)–BRCA pathway is critical for the repair of DNA interstrand crosslinks (ICLs) and the maintenance of chromosome stability. A key step in FA-BRCA pathway activation is the covalent attachment of monoubiquitin to FANCD2 and FANCI. Monoubiquitinated FANCD2 and FANCI localize in chromatin-associated nuclear foci where they interact with several well-characterized DNA repair proteins. Importantly, very little is known about the structure, function, and regulation of FANCD2. Herein, we describe the identification and characterization of a CUE (coupling of ubiquitin conjugation to endoplasmic reticulum degradation) ubiquitin-binding domain (UBD) in FANCD2, and demonstrate that the CUE domain mediates noncovalent binding to ubiquitin in vitro. We show that although mutation of the CUE domain destabilizes FANCD2, the protein remains competent for DNA damage-inducible monoubiquitination and phosphorylation. Importantly, we demonstrate that the CUE domain is required for interaction with FANCI, retention of monoubiquitinated FANCD2, and FANCI in chromatin, and for efficient ICL repair. Our results suggest a model by which heterodimerization of monoubiquitinated FANCD2 and FANCI in chromatin is mediated in part through a noncovalent interaction between the FANCD2 CUE domain and monoubiquitin covalently attached to FANCI, and that this interaction shields monoubiquitinated FANCD2 from polyubiquitination and proteasomal degradation.
Introduction
Fanconi anemia (FA) is a rare, recessive disease characterized by congenital abnormalities, bone marrow failure, hematologic malignancies, and elevated cancer risk.1 FA is caused by biallelic mutation in any 1 of the following 15 genes: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ/BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4. The FA proteins together with BRCA1 function cooperatively in the FA-BRCA pathway to repair damaged DNA and prevent cellular transformation.2 Disruption of the FA-BRCA pathway leads to cellular hypersensitivity to the cytotoxic and clastogenic effects of DNA interstrand crosslinking agents.3
The FA-BRCA pathway is activated after exposure to DNA damaging agents and during S-phase of the cell cycle.4,5 Activation of the pathway occurs when the core FA complex composed of FANCA, B, C, E, F, G, L, and M, and other proteins, assembles in the nucleus and monoubiquitinates the paralogous proteins FANCD2 and FANCI.4,6,7 FANCL, a RING domain-containing protein is the catalytic E3 ubiquitin ligase subunit of the FA core complex, whereas UBE2T is the E2 conjugating enzyme.8-10 Monoubiquitination of FANCD2 and FANCI targets these proteins to discrete chromatin-associated nuclear foci, where they interact with several key DNA repair proteins, including BRCA1, FANCD1/BRCA2, and RAD51.4,5,11 After repair, FANCD2 and FANCI are deubiquitinated by the USP1/UAF1 complex facilitating the release of these proteins from chromatin.12,13 Recent studies indicate that FANCD2 monoubiquitination is necessary for the recruitment of the FAN1 and SLX4/FANCP endonucleases to sites of DNA damage.14-19 Despite the critical functions of FANCD2 and FANCI in interstrand crosslink (ICL) repair, very little is known about their structure, function, and regulation.
Ubiquitin signaling is essential for a vast array of cellular processes including protein trafficking, histone modification, transcriptional regulation, as well as DNA repair.20 One way in which the ubiquitin signal is recognized and interpreted is through noncovalent interaction with proteins harboring ubiquitin-binding domains (UBDs). Several types of UBDs exist including the ubiquitin-associated motif, ubiquitin-interacting motif, ubiquitin-binding zinc finger domain (UBZ), and the CUE domain (for coupling of ubiquitin conjugation to endoplasmic reticulum degradation).21 The CUE domain is a 42 to 43 amino acid sequence identified by its similarity to a region of the yeast Cue1 protein.22,23 Importantly, several proteins harboring UBDs are themselves monoubiquitinated. The structural arrangement of a UBD and a monoubiquitinated residue residing on the same protein has the potential to promote noncovalent intra or intermolecular interactions. Indeed, coupled monoubiquitination has recently been demonstrated to play a key role in the regulation of the translesion DNA synthesis (TLS) polymerase pol η.24
As FANCD2 is known to be monoubiquitinated in response to DNA damage,4 we have long suspected that FANCD2 harbors a UBD. In this study we describe the identification of a CUE UBD in the amino-terminus of FANCD2 and demonstrate that this domain mediates noncovalent interaction between FANCD2 and ubiquitin. We demonstrate that mutation of the CUE domain leads to a decrease in protein stability that can be rescued by inhibition of the proteasome. Multiple distinct FANCD2 CUE mutants retain the ability to undergo DNA damage-inducible monoubiquitination and phosphorylation indicating that reduced protein stability is not a general consequence of protein misfolding. Importantly, we establish that the CUE domain is required for efficient interaction between FANCD2 and FANCI and is essential for the localization of monoubiquitinated FANCD2 and FANCI in chromatin. Consequently, FANCD2 CUE mutants fail to correct the mitomycin C (MMC) hypersensitivity of FA-D2 (FANCD2−/−) patient cells. Taken together, our results suggest that the heterodimerization of monoubiquitinated FANCD2 and FANCI in chromatin is mediated in part through a noncovalent interaction between the FANCD2 CUE domain and monoubiquitin covalently linked to K523 of FANCI, and that this interaction shields monoubiquitinated FANCD2 from polyubiquitination and proteasomal degradation.
Methods
Cell culture and antibodies
PD20 (FA-D2 [FANCD2−/−]), HeLa and COS-7 cells were grown in DMEM media supplemented with 12% v/v fetal bovine serum, l-glutamine, and penicillin/streptomycin. Stable FA-D2 cells were generated by infection with pMMP Moloney murine leukemia or pLenti6.2/V5-DEST (Invitrogen) virus harboring wild-type (WT) or mutant FANCD2 cDNAs.4,25 Stable cell lines were grown in Dulbecco modified Eagle medium (DMEM) supplemented with either 1 μg/mL puromycin or 2 μg/mL blasticidin. The following antibodies were used: rabbit polyclonal antisera against FANCD2 (NB100-182; Novus Biologicals), FANCI (Dr Patrick Sung, Yale University and A300-212A; Bethyl Laboratories), H2A (07-146; Millipore), and mouse monoclonal antisera against α-tubulin (MS-581-PO; Lab Vision) and V5 (R96025; Invitrogen).
Immunofluorescence microscopy
For immunofluorescence microscopy (IF) freely soluble cellular proteins were pre-extracted with 0.3% v/v Triton X-100 and cells fixed in 4% w/v paraformaldehyde and 2% w/v sucrose at 4°C followed by permeabilization in 0.3% v/v Triton X-100 in phosphate-buffered saline (PBS). Fixed cells were blocked for 30 minutes in antibody dilution buffer (5% v/v goat serum, 0.1% v/v NP-40, in PBS) and incubated with primary antibody for 1 hour. Cells were washed 3 times in PBS and incubated for 30 minutes at room temperature with an Alexa fluor 488–conjugated secondary antibody. Nuclear foci were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software.
Immunoprecipitation
Cells were lysed in NETN100 (20mM Tris-HCl pH 7.4, 0.1% v/v NP-40, 100mM NaCl, 1mM EDTA, 1mM Na3O4V, 1mM NaF, supplemented with protease inhibitors), incubated on ice and sonicated briefly. Whole-cell lysates (WCLs; 800 μg) were incubated with 3 μg of antibodies against FANCD2 (FI-17; Santa Cruz), V5 (R96025; Invitrogen), or mouse immunoglobulin (IgG; 12-371B; Millipore).
Plasmids and site-directed mutagenesis
The FANCD2-P204A, -LP215AA, and -LL234AA cDNAs were generated by site-directed mutagenesis of the WT FANCD2 cDNA using the Quikchange site-directed mutagenesis kit (Stratagene). The forward and reverse oligonucleotide sequences used are as follows: P204A FP, 5′-GATCATGCAGCTGATCAGTATTGCTGCAGAGAACCTGCAGCATGACATCAT-3′; P204A RP, 5′-ATGATGTCATGCTGCAGGTTCTCTGCAGCAATACTGATCAGCTGCATGATC-3′; LP2155AA FP, 5′-GAGAACCTGCAGCATGACATCATCACCAGCGCAGCTGAGATCCTAGGGGATTCCCAGCACGCTGAT-3′; LP215AA RP, 5′-ATCAGCGTGCTGGGAATCCCCTAGGATCTCAGCTGCGCTGGTGATGATGTCATGCTGCAGGTTCTC-3′; LL234AA FP, 5′-CACGCTGATGTGGGGAAAGAACTCAGTGACGCAGCGATAGAGAATACTTCACTCACTGTCCCAATC-3′; LL234AA RP GATTGGGACAGTGAGTGAAGTATTCTCTATCGCTGCGTCACTGAGTTCTTTCCCCACATCAGCGTG. The mammalian expression vector pDEST40 (Invitrogen) and the bacterial expression vector pDEST49 (Invitrogen), allow for the expression of C-terminal H6/V5 fusion proteins. pGEX-2TK (GE Healthcare) was used for the expression of carboxy-terminus V5/6xHistidine (H6) fusion proteins.
Protein purification and ubiquitin-binding assays
V5/H6 fusion protein expression in Escherichia coli BL21 (DE3; Stratagene) was induced with 0.2% L-arabinose for 10 hours at room temperature. Cell pellets were collected, resuspended in ice-cold lysis buffer (50mM potassium phosphate, pH 7.8, 400mM NaCl, 10mM KCl, 10% v/v glycerol, 0.5% v/v Triton-X-100, 10mM imidazole) with sonication. V5/H6 fusion proteins were purified with Ni-NTA agarose (Invitrogen) according to the manufacturer's instructions. GST fusion protein expression was induced with 400μM IPTG for 5 hours at 30°C. Cell pellets were collected, resuspended in ice-cold lysis buffer (50mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 150mM NaCl, 1mM EDTA, 1mM EGTA [ethylene glyco-bis(b-aminoethyl ester)-N,N,N′,N′-tetraacetic acid], 10% v/v glycerol, 1% v/v Triton-X-100, 25mM NaF, 10μM ZnCl2, pH 7.5) containing protease inhibitors (Roche), sonicated and centrifuged for 15 minutes at 13 400g. Purified proteins were resolved on NuPAGE 4%-12% Bis-Tris gels (Invitrogen) and protein purity assessed via Coomassie staining. For ubiquitin-binding assays between 4 and 40 ng of purified protein, or 5 mg of cleared bacterial WCLs, was incubated with ubiquitin sepharose (Boston Biochem) in lysis buffer for at least 1 hour at 4°C. Beads were pelleted via brief centrifugation and washed 3 times in lysis buffer. After the final wash, beads were heated in 2× NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) and proteins resolved on NuPAGE 4%-12% Bis-Tris gels.
DNA damage assays
MMC survival assays and FACS analysis were carried out as previously described.26,27 For chromosome breakage assays, cells were treated with MMC for 24 hours and harvested for chromosome preparations using standard conditions as previously described.28 Metaphase chromosomes were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software.
Cellular fractionation and λ-phosphatase assay
Soluble proteins were removed by extraction in cytoskeletal buffer (CSK; 10mM PIPES pH 6.8, 300mM sucrose, 100mM NaCl, 3mM MgCl2, 1mM EGTA, and 0.5% v/v Triton-X-100) for 10 minutes at 4°C. Pellets were washed once with CSK buffer, lysed in sodium dodecyl sulfate (SDS) sample buffer (2% w/v SDS, 50mM Tris-HCl pH 7.4, 10mM, and EDTA), boiled for 15 minutes and sonicated. The λ-phosphatase assay was preformed as previously described.29
Results
FANCD2 contains a highly conserved putative CUE ubiquitin-binding domain
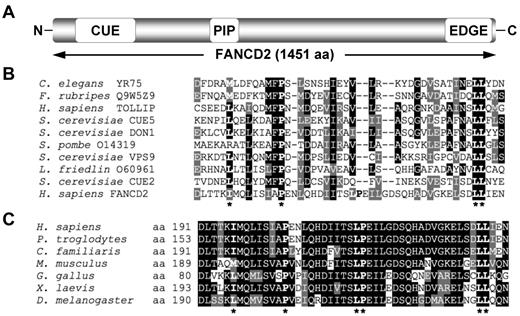
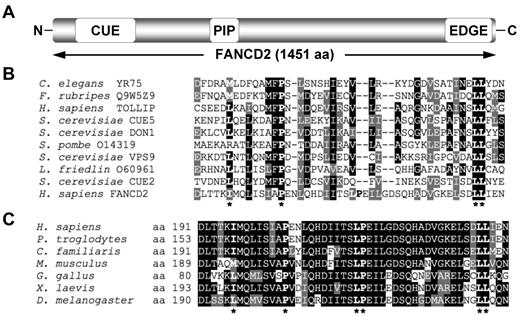
To gain greater insight into the domain organization and structure of the FANCD2 protein, the 1451-amino acid sequence of FANCD2 was fragmented into ∼ 150-amino acid segments and analyzed using Pfam and SMART databases. This approach yielded a low-homology hit between an amino-terminus fragment of FANCD2 and the CUE domain of the Saccharomyces cerevisiae Vps9 protein (Figure 1A). A sequence alignment of this amino-terminus fragment of FANCD2 with several known CUE domains revealed high conservation of a proline residue and dileucine motif, characteristic of CUE UBDs, as well as a number of other hydrophobic residues thought to be important for noncovalent interaction with ubiquitin (Figure 1B). A sequence alignment of this region of FANCD2 among vertebrates demonstrates strong evolutionary conservation (Figure 1C). Using PORTER (http://distill.ucd.ie/porter/) secondary protein structure prediction software,30 the putative CUE domain was predicted to contain 3 consecutive α-helices, with the conserved proline and dileucine residues residing in helix 1 and 3, respectively. Importantly, this arrangement is common among several CUE domains including that of the yeast Cue2 protein.31 Recently, the crystal structure of the murine Fancd2-Fanci (ID) complex was solved.32 In support of our secondary structure prediction, the crystallized ID structure demonstrates that the Fancd2 CUE domain does adopt a triple α-helical arrangement (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Moreover, the CUE domain of Fancd2 is in close proximity to K522 of Fanci (equivalent residue to K523 of human FANCI; supplemental Figure 1B), the residue that undergoes monoubiquitination, suggesting that the heterodimerization of FANCD2 and FANCI may be mediated by an interaction between the CUE domain of FANCD2 and ubiquitin covalently linked to FANCI K523.
FANCD2 contains a putative CUE ubiquitin-binding domain. (A) Schematic of the FANCD2 protein indicating the amino terminus CUE domain, the PCNA-interaction motif (PIP), and the carboxy terminus EDGE motif. (B) A ClustalW alignment of the amino terminus of FANCD2 with several known CUE domains. (C) A ClustalW alignment of the amino terminus of FANCD2 from several species demonstrates high conservation of this region. Asterisks indicate highly conserved amino acid residues known to be important for noncovalent interaction with ubiquitin.
FANCD2 contains a putative CUE ubiquitin-binding domain. (A) Schematic of the FANCD2 protein indicating the amino terminus CUE domain, the PCNA-interaction motif (PIP), and the carboxy terminus EDGE motif. (B) A ClustalW alignment of the amino terminus of FANCD2 with several known CUE domains. (C) A ClustalW alignment of the amino terminus of FANCD2 from several species demonstrates high conservation of this region. Asterisks indicate highly conserved amino acid residues known to be important for noncovalent interaction with ubiquitin.
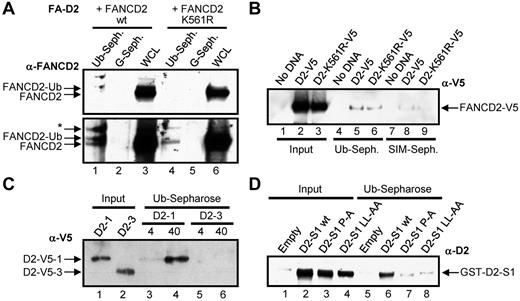
FANCD2 interacts with ubiquitin noncovalently
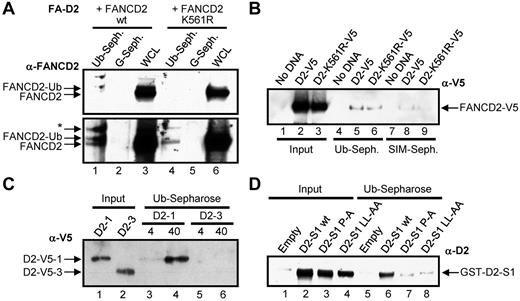
To determine whether FANCD2 does indeed interact noncovalently with ubiquitin, WCLs from FA-D2 (FANCD2−/−) patient cells reconstituted with either WT FANCD2 or FANCD2 K561R were incubated with ubiquitin or protein G–conjugated sepharose. Both nonubiquitinated and monoubiquitinated FANCD2 noncovalently bound to ubiquitin but not to protein G (Figure 2A bottom panel lanes 1 and 4). In another approach, WCLs from HeLa cells transiently expressing V5-tagged WT FANCD2 or FANCD2 K561R were incubated with ubiquitin-conjugated sepharose or SUMO-interacting motif (SIM)–conjugated sepharose, as a negative control. Once again FANCD2-V5 and FANCD2-K561R-V5 noncovalently bound to ubiquitin (Figure 2B lanes 5-6). A weaker lower affinity interaction between FANCD2 and SIM was also detected (Figure 2B lanes 8-9; supplemental Figure 2A). Next we bacterially purified 2 FANCD2 fragments, FANCD2-1, encompassing amino acids 1-254 harboring the CUE domain, and FANCD2-3, encompassing amino acids 585-819, and incubated them with ubiquitin-sepharose. FANCD2-1 noncovalently bound to ubiquitin in a concentration-dependent manner, whereas FANCD2-3 did not (Figure 2C lanes 3-6). Finally, we incubated bacterial lysates expressing GST-tagged FANCD2 solenoid 1 (S1)32 encompassing the CUE domain, as well as S1 harboring P204A and LL234AA missense mutations in the CUE domain (see the next section). WT FANCD2 S1 bound efficiently to ubiquitin, whereas ubiquitin binding was significantly compromised for both FANCD2 S1 CUE mutants, even taking into account the slightly reduced levels of expression of the CUE mutants (Figure 2D, supplemental Figure 2B). Taken together, these results demonstrate that FANCD2 can noncovalently interact with ubiquitin and that this interaction is mediated by the amino-terminus CUE domain. It is important to note that based on the absence of M and F residues preceding the conserved P, it is probable that the FANCD2 CUE domain represents a low affinity UBD similar to that of S cerevisiae Cue1p, and unlike that of S cerevisiae Vps9p (Figure 1A).23
FANCD2 noncovalently interacts with ubiquitin. (A) WCLs from FA-D2 cells expressing WT or mutant FANCD2 were incubated with ubiquitin or protein G-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against FANCD2. * indicates nonspecific band; WCL, whole-cell lysate; Ub-seph., ubiquitin-conjugated sepharose; and G-seph., protein G-conjugated sepharose. (B) WCLs from HeLa cells expressing WT or mutant FANCD2-V5 were incubated with ubiquitin or SIM-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against V5. (C) Bacterially purified fragments of FANCD2 were incubated with ubiquitin-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against V5. (D) Bacterial WCLs expressing GST-Empty or GST-tagged WT FANCD2 solenoid 1 (D2-S1 WT), FANCD2 S1 P204A (D2-S1 P-A), or FANCD2 S1 LL234AA (D2-S1 LL-AA) were incubated with ubiquitin-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against FANCD2.
FANCD2 noncovalently interacts with ubiquitin. (A) WCLs from FA-D2 cells expressing WT or mutant FANCD2 were incubated with ubiquitin or protein G-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against FANCD2. * indicates nonspecific band; WCL, whole-cell lysate; Ub-seph., ubiquitin-conjugated sepharose; and G-seph., protein G-conjugated sepharose. (B) WCLs from HeLa cells expressing WT or mutant FANCD2-V5 were incubated with ubiquitin or SIM-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against V5. (C) Bacterially purified fragments of FANCD2 were incubated with ubiquitin-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against V5. (D) Bacterial WCLs expressing GST-Empty or GST-tagged WT FANCD2 solenoid 1 (D2-S1 WT), FANCD2 S1 P204A (D2-S1 P-A), or FANCD2 S1 LL234AA (D2-S1 LL-AA) were incubated with ubiquitin-conjugated sepharose and bound proteins resolved and immunoblotted with antibodies against FANCD2.
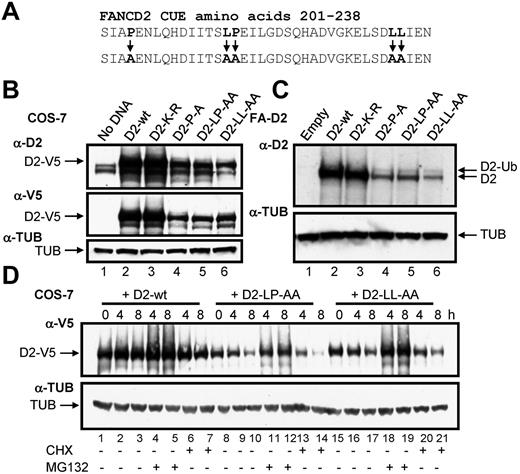
Mutation of the CUE domain destabilizes FANCD2
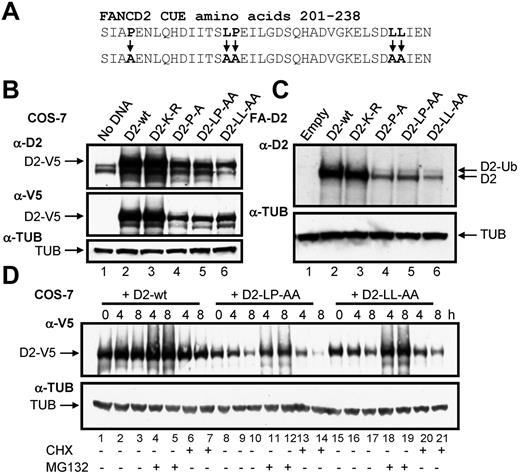
To determine the function of the CUE domain we used a site-directed mutagenesis approach to generate mammalian expression constructs of the FANCD2 CUE missense mutants P204A and LL234AA (Figure 3A). Previous studies demonstrated that mutation of the conserved proline and dileucine residues impairs the ability of the CUE domain to noncovalently interact with ubiquitin.23 In addition, we generated a LP215AA CUE mutant as these residues also demonstrated a high degree of evolutionary conservation (Figure 1C). PORTER secondary structure prediction analysis revealed that the triple helical supersecondary structure of the CUE domain should not be disrupted by these mutations. Equal quantities of WT and CUE mutant FANCD2-V5 constructs were transfected into COS-7 cells and protein expression was assessed. Intriguingly, protein levels of all 3 FANCD2 CUE mutants were significantly reduced compared with that of WT FANCD2 (Figure 3B, supplemental Figure 3). We also generated FA-D2 patient cell lines stably expressing WT FANCD2 or the FANCD2 missense mutants K561R, P204A, LP215AA, and LL234AA. Although both WT FANCD2 and the K561R mutant displayed robust levels of expression, expression levels of all 3 CUE mutants were again reduced (Figure 3C). Reduced protein expression was consistently observed for multiple independently generated FA-D2 CUE mutant lines (results not shown). However, the FANCD2 CUE mutants remained competent for monoubiquitination indicating that protein folding and overall tertiary structure was not overtly affected. To determine whether reduced protein stability was a consequence of increased protein turnover via proteasome-mediated degradation, COS-7 cells transiently expressing WT FANCD2-V5 or the CUE mutants were treated with the proteasome inhibitor MG132 and protein levels measured. Although MG132 treatment had little effect on WT protein levels, a significant increase in FANCD2 LP215AA protein levels was observed (Figure 3D lanes 11-12). Similar results were obtained for both FANCD2 P204A and LL234AA (Figure 3D lanes 18-19 and results not shown). Furthermore, treatment with the protein translation inhibitor cycloheximide led to a rapid and pronounced reduction in FANCD2 LP214AA protein levels (Figure 3D lane 14). Moreover, deletion of the entire CUE domain (amino acids 1-254) led to a marked destabilization of the protein (supplemental Figure 4). Collectively, these results suggest that the FANCD2 CUE domain is a major determinant of protein stability and may protect FANCD2 from rapid proteasome-mediated turnover.
Mutation of the CUE domain impairs FANCD2 protein stability. (A) Using site-directed mutagenesis, 3 CUE domain missense mutations were generated. (B) WCLs from COS-7 cells expressing WT or mutant FANCD2 were resolved and immunoblotted with antibodies against FANCD2, V5, and α-tubulin (TUB). (C) WCLs from FA-D2 cells reconstituted with WT or mutant FANCD2 were resolved and immunoblotted with antibodies against FANCD2 and tubulin. h indicates hours. (D) COS-7 cells were transfected with WT or mutant FANCD2. Forty-eight hours later cells were untreated or treated with 10μM MG132 or 35μM cyclohexamide (CHX). Pellets were collected at the indicated times and WCLs generated. Proteins were resolved and immunoblotted with antibodies against V5 and α-tubulin.
Mutation of the CUE domain impairs FANCD2 protein stability. (A) Using site-directed mutagenesis, 3 CUE domain missense mutations were generated. (B) WCLs from COS-7 cells expressing WT or mutant FANCD2 were resolved and immunoblotted with antibodies against FANCD2, V5, and α-tubulin (TUB). (C) WCLs from FA-D2 cells reconstituted with WT or mutant FANCD2 were resolved and immunoblotted with antibodies against FANCD2 and tubulin. h indicates hours. (D) COS-7 cells were transfected with WT or mutant FANCD2. Forty-eight hours later cells were untreated or treated with 10μM MG132 or 35μM cyclohexamide (CHX). Pellets were collected at the indicated times and WCLs generated. Proteins were resolved and immunoblotted with antibodies against V5 and α-tubulin.
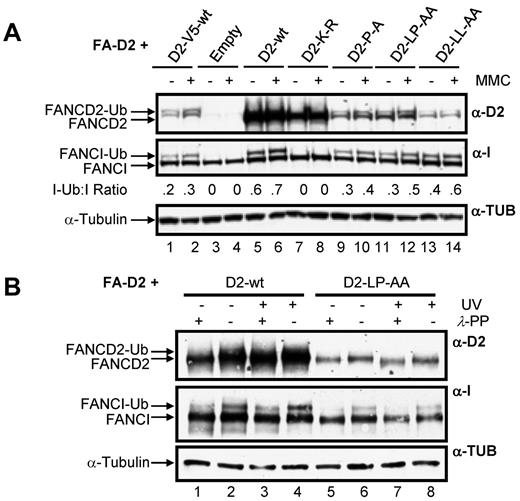
Disruption of the FANCD2 CUE domain does not impair DNA damage-inducible FANCD2 and FANCI posttranslational modification
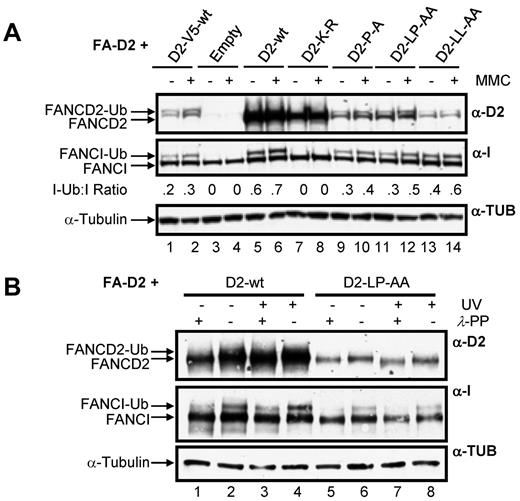
A key step in the activation of the FA-BRCA pathway is the monoubiquitination of FANCD2 and FANCI. Therefore, we next examined the effect of mutation of the CUE domain on this important posttranslational modification. For this experiment, and all subsequent experiments, to control for the effects of the large difference in WT FANCD2 and CUE mutant protein expression levels, we included FA-D2 cells stably expressing WT FANCD2-V5. Importantly, WT FANCD2-V5 and the FANCD2 CUE mutants are expressed at similar levels in the FA-D2 cells (Figure 4A compare lanes 1-2 to lanes 9-14). Moreover, WT FANCD2-V5 undergoes DNA damage-inducible monoubiquitination and corrects the MMC-hypersensitivity of FA-D2 patient cells (see Figure 7). All FANCD2 CUE mutants remained competent for both spontaneous and MMC-inducible monoubiquitination (Figure 4A lanes 9-14). Previous studies have shown that FANCD2 and FANCI monoubiquitination are interdependent.6,7 Consistent with these studies, mutation of FANCD2 K561 resulted in complete abrogation of FANCI monoubiquitination (Figure 4A lanes 7 and 8). In contrast, mutation of the CUE domain resulted in a modest reduction in MMC-inducible FANCI monoubiquitination compared with cells expressing WT FANCD2 (Figure 4A compare lanes 5-6 to lanes 9-14). However, similar levels of FANCI monoubiquitination were observed in FA-D2 cells expressing WT FANCD2-V5 (Figure 4A compare lanes 1-2 to lanes 9-14), indicating that attenuated FANCI monoubiquitination was a consequence of reduced FANCD2 protein expression, and not mutation of the CUE domain per se. We also examined the ability of the FANCD2 CUE mutants to undergo DNA damage-inducible phosphorylation by comparing electrophoretic mobility in the absence and presence of λ-phosphatase.29 Mutation of the CUE domain had no discernible effect on DNA damage-inducible FANCD2 phosphorylation, as demonstrated by an increase in protein mobility after treatment of WCLs with λ-phosphatase (Figure 4B compare lanes 7 and 8). Taken together these findings demonstrate that although mutation of the FANCD2 CUE domain leads to a pronounced decrease in protein stability, tertiary protein structure is not overtly impacted by mutation of this domain.
Disruption of the FANCD2 CUE domain does not impair DNA damage-inducible FANCD2 and FANCI posttranslational modification. (A) FA-D2 cells were untreated or treated with 250nM MMC. Twelve hours later pellets were collected and lysed and proteins resolved and immunoblotted with antibodies against FANCD2, FANCI, and tubulin. (B) FA-D2 cells were untreated or treated with 20 J/m2 UV-C irradiation. Six hours later pellets were collected, and WCLs were generated. For each sample, an aliquot of the WCLs was then treated with λ-phosphatase (λ-PP). Proteins were resolved and immunoblotted with antibodies against FANCD2, FANCI, and α-tubulin.
Disruption of the FANCD2 CUE domain does not impair DNA damage-inducible FANCD2 and FANCI posttranslational modification. (A) FA-D2 cells were untreated or treated with 250nM MMC. Twelve hours later pellets were collected and lysed and proteins resolved and immunoblotted with antibodies against FANCD2, FANCI, and tubulin. (B) FA-D2 cells were untreated or treated with 20 J/m2 UV-C irradiation. Six hours later pellets were collected, and WCLs were generated. For each sample, an aliquot of the WCLs was then treated with λ-phosphatase (λ-PP). Proteins were resolved and immunoblotted with antibodies against FANCD2, FANCI, and α-tubulin.
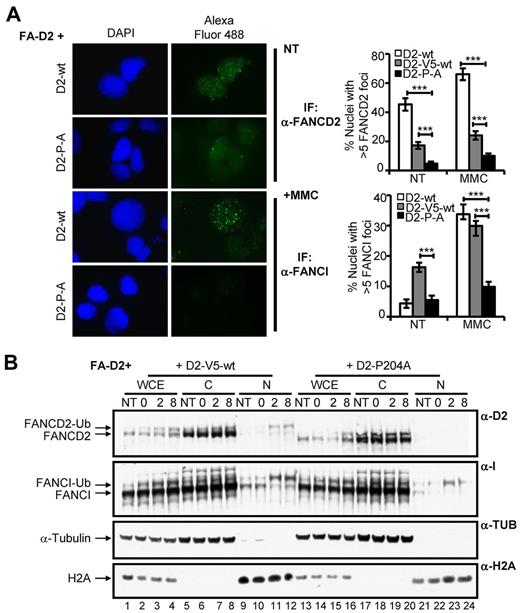
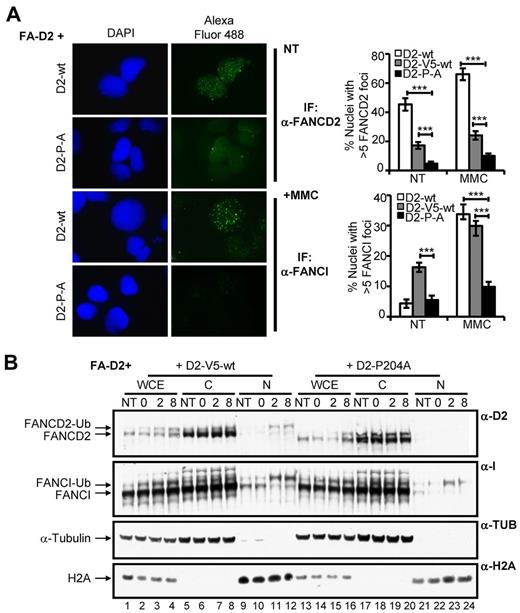
The CUE domain is required for FANCD2 and FANCI nuclear foci formation and chromatin localization
After exposure to DNA damaging agents, FANCD2 and FANCI assemble in discrete chromatin-associated nuclear foci to facilitate DNA repair.4,6,7 To determine whether the FANCD2 CUE domain is important for FANCD2/I nuclear foci formation, FA-D2 cells stably expressing WT or mutant FANCD2 were treated with MMC and FANCD2/I nuclear foci formation was assessed using IF. Both spontaneous and DNA damage-inducible FANCD2 nuclear foci formation were markedly impaired in cells expressing mutant FANCD2 (Figure 5A, supplemental Figure 5A). For example, after MMC treatment ∼ 66% of cells expressing WT FANCD2 displayed nuclear foci compared with only ∼ 10% of cells expressing FANCD2 P204A (P < .001). FANCI nuclear foci formation was also markedly impaired in FA-D2 cells reconstituted with the CUE mutants. For example, after exposure to MMC only approximately 10% of FA-D2 cells expressing FANCD2 P204A were positive for FANCI nuclear foci formation, in comparison with ∼ 33% of cells expressing WT FANCD2 (P < .001; Figure 5A, supplemental Figure 5B). Importantly, reduced FANCD2 and FANCI nuclear foci formation was not solely a consequence of decreased levels of expression of the CUE mutants as foci formation was also significantly reduced compared with FA-D2 cells stably expressing similar levels of WT FANCD2-V5 (Figure 5A, supplemental Figure 5A-B). We next performed a chromatin fractionation experiment with FA-D2 cells stably expressing WT FANCD2-V5 or FANCD2 P204A to corroborate our IF findings. Cells were incubated in the absence or presence of MMC for 12 hours, washed to remove the drug and allowed to recover in drug-free media. In agreement with our IF findings, despite similar total levels of FANCD2, we observed greatly reduced levels of monoubiquitinated FANCD2 and FANCI in chromatin fractions of FA-D2 cells expressing FANCD2 P204A, compared with cells expressing WT FANCD2-V5 (Figure 5B compare lanes 23-24 with lanes 11-12). Taken together, our results demonstrate that the FANCD2 CUE domain is required for the localization and/or stabilization of FANCD2 and FANCI in chromatin.
The CUE domain is required for FANCD2 and FANCI nuclear foci formation and chromatin localization. (A) FA-D2 cells reconstituted with WT or mutant FANCD2 were untreated or treated with 250nM MMC for 12 hours and then fixed and immunostained with antibodies against FANCD2 and FANCI. Nuclear foci were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software. The percentage of nuclei with greater than 5 foci were scored and plotted in the indicated histograms (***P < .001). (B) FA-D2 cells reconstituted with WT or mutant FANCD2 were untreated or treated with 250nM MMC for 12 hours, released from treatment and pellets collected at the indicated time points. Pellets were divided and one-half was lysed in 2% SDS lysis buffer, sonicated, and boiled (WCE). The remaining pellet was sequentially lysed in cytoskeletal buffer to extract soluble proteins (C) and then 2% SDS lysis buffer with sonication and boiling to extract proteins bound to chromatin (N). Proteins were resolved and immunoblotted with antibodies against FANCD2, FANCI, α-tubulin, and H2A.
The CUE domain is required for FANCD2 and FANCI nuclear foci formation and chromatin localization. (A) FA-D2 cells reconstituted with WT or mutant FANCD2 were untreated or treated with 250nM MMC for 12 hours and then fixed and immunostained with antibodies against FANCD2 and FANCI. Nuclear foci were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software. The percentage of nuclei with greater than 5 foci were scored and plotted in the indicated histograms (***P < .001). (B) FA-D2 cells reconstituted with WT or mutant FANCD2 were untreated or treated with 250nM MMC for 12 hours, released from treatment and pellets collected at the indicated time points. Pellets were divided and one-half was lysed in 2% SDS lysis buffer, sonicated, and boiled (WCE). The remaining pellet was sequentially lysed in cytoskeletal buffer to extract soluble proteins (C) and then 2% SDS lysis buffer with sonication and boiling to extract proteins bound to chromatin (N). Proteins were resolved and immunoblotted with antibodies against FANCD2, FANCI, α-tubulin, and H2A.
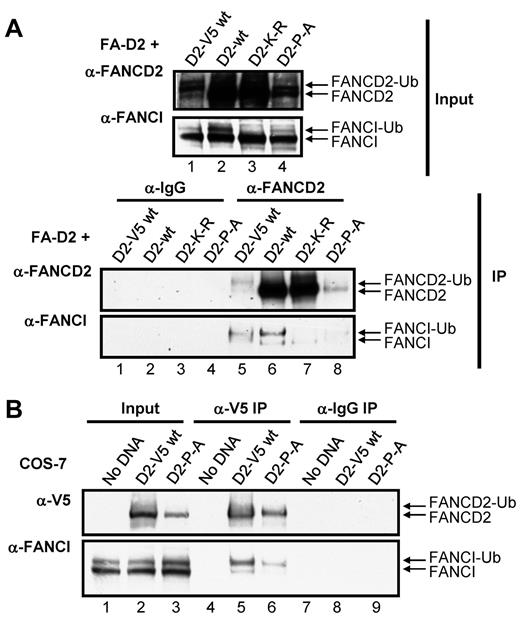
The FANCD2 CUE domain is required for efficient interaction with FANCI
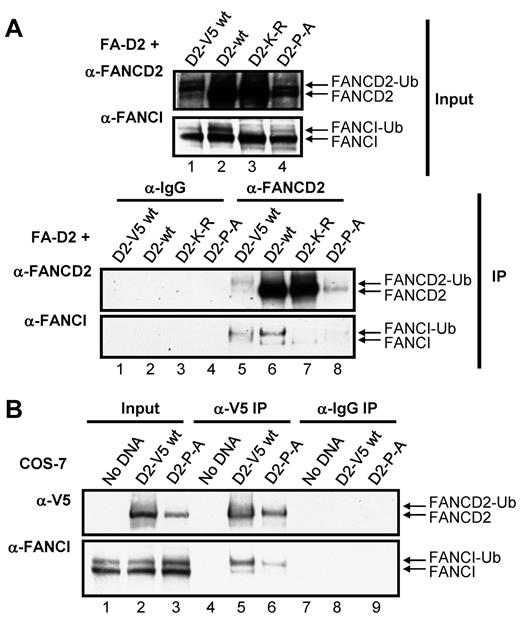
Several reports have suggested that the stability of FANCD2 and FANCI are interdependent and that FANCD2/I monoubiquitination may act to stabilize the FANCI-FANCD2 (ID) complex.7,32 We hypothesized that mutation of the CUE domain might impair ID complex formation. To test this hypothesis, we examined the effect of mutation of the CUE domain on the ability of FANCD2 and FANCI to coimmunoprecipitate. FANCD2 immune complexes immunoprecipitated from FA-D2 cells reconstituted with WT or mutant FANCD2 were examined for the presence of FANCI. WT FANCD2 immunoprecipitated the highest levels of both nonubiquitinated and monoubiquitinated FANCI (Figure 6A IP: α-FANCI panel lane 6). These results were not surprising given the robust level of FANCD2 expression in these cells (Figure 6A input: α-FANCD2 panel lane 2). To gain a more meaningful understanding of the effect of CUE mutation on the FANCD2-FANCI interaction, we also included FA-D2 cells stably reconstituted with WT FANCD2-V5, as WT FANCD2-V5 and FANCD2-P204A are expressed at similar levels (Figure 6A input: α-FANCD2 panel compare lanes 1 and 4) therefore, any difference in the ability of these proteins to immunoprecipitate FANCI is probably solely a consequence of CUE mutation and not an effect of protein expression. Importantly, WT FANCD2-V5 and FANCD2-P204A immunoprecipitated similar levels of FANCD2 immune complexes (Figure 6A IP:α-FANCD2 panel compare lanes 5 and 8). However, in contrast, WT FANCD2-V5 immunoprecipitated considerably higher levels of FANCI compared with FANCD2-P204A (Figure 6A IP:α-FANCI panel compare lanes 5 and 8). Interestingly, with the exception of the K561R mutant, monoubiquitinated FANCI was the predominant form immunoprecipitated, supporting previous studies suggesting that FANCD2/I monoubiquitination may stabilize ID complex formation.32 In an alternative approach, we transiently transfected COS-7 cells with WT FANCD2-V5 or FANCD2-P204A-V5 and examined the association with endogenous FANCI. Despite lower expression levels of the FANCD2-P204A-V5 mutant compared with WT FANCD2-V5, similar levels of FANCD2 immune complexes were precipitated (Figure 6B α-V5 panel compare lanes 5 and 6). Again, considerably lower levels of FANCI coimmunoprecipitated with FANCD2-P204A-V5 compared with WT FANCD2-V5 (Figure 6B α-FANCI panel compare lanes 5 and 6). Taken together, these results suggest that FANCI-FANCD2 binding may be facilitated, at least in part, by the CUE domain.
The FANCD2 CUE domain is required for efficient interaction with FANCI. (A) WCLs from FA-D2 cells reconstituted with WT or mutant FANCD2 were immunoprecipitated with antibodies against FANCD2 or mouse IgG. Bound proteins were resolved and immunoblotted with antibodies against FANCD2 and FANCI. (B) WCLs from COS-7 cells transfected with WT or mutant FANCD2-V5 were immunoprecipitated with antibodies against V5 or mouse IgG. Bound proteins were resolved and immunoblotted with antibodies against V5 and FANCI.
The FANCD2 CUE domain is required for efficient interaction with FANCI. (A) WCLs from FA-D2 cells reconstituted with WT or mutant FANCD2 were immunoprecipitated with antibodies against FANCD2 or mouse IgG. Bound proteins were resolved and immunoblotted with antibodies against FANCD2 and FANCI. (B) WCLs from COS-7 cells transfected with WT or mutant FANCD2-V5 were immunoprecipitated with antibodies against V5 or mouse IgG. Bound proteins were resolved and immunoblotted with antibodies against V5 and FANCI.
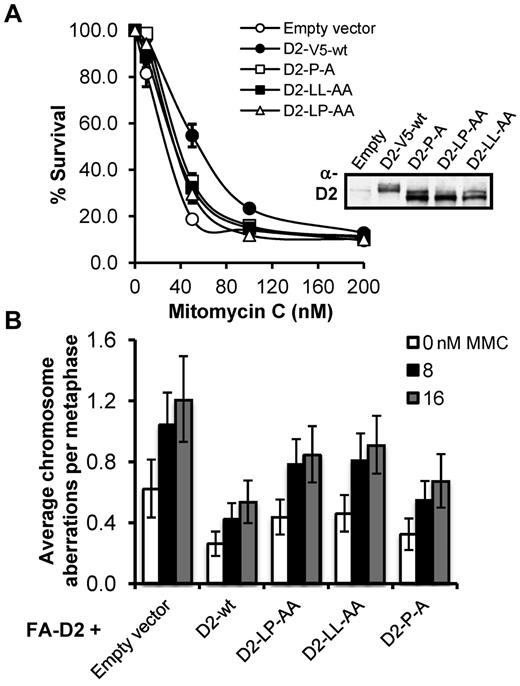
FANCD2 CUE mutants fail to rescue the MMC-hypersensitivity of FA-D2 patient cells
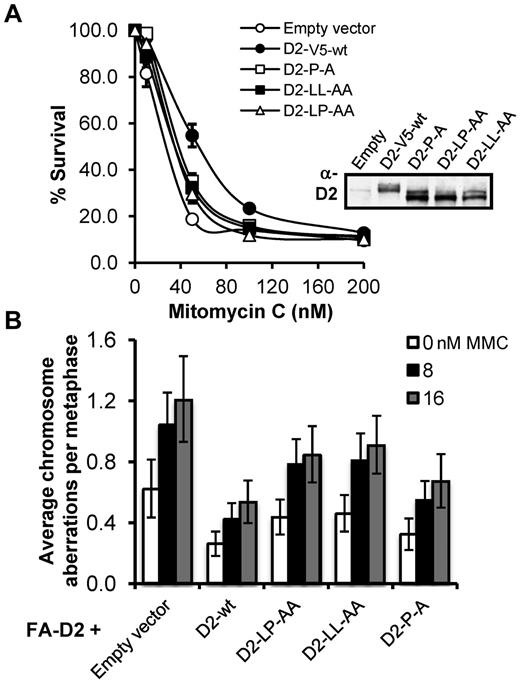
FA patient-derived cells are characteristically hypersensitive to the cytotoxic and clastogenic affects of DNA interstrand crosslinking agents.3 Therefore, we next assessed the ability of the FANCD2 CUE mutants to rescue the MMC hypersensitivity of FA-D2 patient cells. To ensure that FANCD2 expression was not a confounding factor in our functional assays we again used FA-D2 cells stably expressing FANCD2-V5 as our WT control. In a MMC cytotoxicity assay, whereas WT FANCD2-V5 rescued the MMC hypersensitivity of FA-D2 cells, all 3 FA-D2 cell lines expressing the FANCD2 CUE mutants displayed intermediate MMC sensitivity (Figure 7A). For example, an approximate 2-fold (P < .05) reduction in the percentage survival was observed for FA-D2 cells expressing FANCD2 P204A, compared with cells expressing WT FANCD2, after exposure to 50nM MMC (Figure 7A). Similarly, in a MMC clastogenicity assay, FA-D2 cells expressing the CUE mutants displayed intermediate sensitivity to the clastogenic effects of MMC (Figure 7B). For example, FA-D2 cells expressing FANCD2 LL234AA exhibited a ∼ 2-fold (P = .002) increase in MMC-induced chromosome aberrations compared with FA-D2 cells expressing WT FANCD2-V5. Furthermore, FACS analysis revealed an intermediate G2/M accumulation of FA-D2 cells expressing the FANCD2 CUE mutants, compared with FA-D2 cells expressing empty vector or FANCD2 K561R, after exposure to MMC (supplemental Figure 6). Taken together, our results demonstrate that the FANCD2 CUE domain is necessary for efficient ICL repair.
The FANCD2 CUE mutants fail to rescue the MMC-hypersensitivity of FA-D2 patient cells. (A) FA-D2 cells reconstituted with WT or mutant FANCD2 were treated with the indicated concentrations of mitomycin C (MMC) for 7 to 10 days. Cells were fixed and stained with crystal violet and percent survival calculated and plotted. Each measurement was performed in triplicate. The averages for 3 independent experiments were calculated and plotted. (B) FA-D2 cells reconstituted with WT or mutant FANCD2 were incubated in the absence or presence of 8 or 16nM MMC for 24 hours and the numbers of chromosome aberrations including gaps and breaks, dicentrics, and complex chromosome aberrations, including radial formations, were scored. Metaphase spreads were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software. At least 80 metaphases were scored per treatment. Error bars represent the standard error of the mean.
The FANCD2 CUE mutants fail to rescue the MMC-hypersensitivity of FA-D2 patient cells. (A) FA-D2 cells reconstituted with WT or mutant FANCD2 were treated with the indicated concentrations of mitomycin C (MMC) for 7 to 10 days. Cells were fixed and stained with crystal violet and percent survival calculated and plotted. Each measurement was performed in triplicate. The averages for 3 independent experiments were calculated and plotted. (B) FA-D2 cells reconstituted with WT or mutant FANCD2 were incubated in the absence or presence of 8 or 16nM MMC for 24 hours and the numbers of chromosome aberrations including gaps and breaks, dicentrics, and complex chromosome aberrations, including radial formations, were scored. Metaphase spreads were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software. At least 80 metaphases were scored per treatment. Error bars represent the standard error of the mean.
Discussion
In this study, we describe the identification and characterization of a CUE UBD in the amino-terminus of the FANCD2 protein. Despite the clear importance of FANCD2 in the repair of ICLs and the maintenance of chromosome stability, its structure, function, and regulation are quite poorly understood. Indeed, to date only 2 functional motifs in FANCD2 have been identified and characterized, including a PCNA-interaction motif, or PIP box, and a carboxy-terminus EDGE motif (Figure 1A).29,33 Thus, the CUE UBD represents the first bona fide domain to be identified in this important protein. Site-directed mutagenesis of residues predicted to play important roles in noncovalent ubiquitin binding revealed several observations about the importance of this domain. First, all missense CUE mutants examined, as well as a CUE deletion mutant, demonstrated reduced protein stability compared with WT FANCD2 and the monoubiquitination defective K561R mutant. Reduced protein stability is unlikely to be an indirect consequence of protein misfolding as all the CUE mutants tested retained the ability to undergo DNA damage-inducible FANCD2 monoubiquitination and phosphorylation, suggesting that their overall structural conformation remained intact. Inhibition of the proteasome partially rescued the stability of the CUE mutants suggesting that an inability to noncovalently interact with ubiquitin may lead to ubiquitin-mediated proteolytic degradation of FANCD2. Second, mutation of the CUE domain reduced the ability of FANCD2 to interact with FANCI, suggesting that ID heterodimerization is facilitated at least in part by the CUE domain. Third, mutation of the CUE domain led to a dramatic reduction in the ability of both FANCD2 and FANCI to assemble into discrete nuclear foci and to localize to chromatin, suggesting that ID complex formation is necessary for retention of both proteins in chromatin. Consequently, the CUE mutants failed to correct the MMC-hypersensitivity of FA-D2 patient cells underscoring the overall functional significance of this UBD. Underscoring the importance of this domain, several FA-D2 patient-derived mutations have been mapped to this region.34
The existence of a CUE UBD in FANCD2 gives rise to at least 3 plausible predictions regarding the behavior of FANCD2; (1) monoubiquitination of FANCD2 causes it to adopt a closed conformation whereby monoubiquitin covalently linked to K561 noncovalently interacts with the CUE domain on the same monomer, (2) monoubiquitination of FANCD2 leads to the formation of a FANCD2 homodimer facilitated through noncovalent interaction between the CUE domain and monoubiquitin covalently linked to K561 on an adjacent monomer, or (3) monoubiquitination of both FANCD2 and FANCI stimulates ID heterodimerization, facilitated in part by noncovalent interaction between the CUE domain and monoubiquitin covalently linked to FANCI K523 (supplemental Figure 7). We favor the latter prediction for several reasons. For example, previous studies demonstrated that monoubiquitinated FANCD2 and FANCI colocalize in discrete nuclear foci and physically interact.6,7 Here, we demonstrated that mutation of the CUE domain reduces the ability of FANCD2 to interact with FANCI, most probably leading to destabilization of the ID complex. A partial disruption of ID complex formation may explain the intermediate ICL sensitivity phenotype observed for FA-D2 cells reconstituted with FANCD2 CUE missense mutants. In addition, as previously mentioned, the crystal structure of the nonubiquitinated murine ID complex was recently solved.32 The dimensions of the tunnels and entrances of the ID structure are sufficiently large to accommodate ubiquitin and suggest that ubiquitin contacts both its conjugated protein and its heterodimerization partner. Importantly, the authors note that the nonubiquitinated ID complex has a short half-life, and speculate that monoubiquitination may enhance ID complex stability.32 Our results are in agreement with this hypothesis and demonstrate that the FANCD2 CUE domain plays a significant role in this endeavor. Based on this model, one would speculate that FANCI also harbors a UBD and that an interaction with ubiquitin conjugated to FANCD2 K561 might also contribute to ID heterodimerization and stabilization. This could explain why mutation of the CUE domain only partially disrupts the interaction with FANCI; perhaps mutation of both UBDs is necessary to completely abolish ID complex formation. Although this is an attractive hypothesis, to date no UBDs have been uncovered in the FANCI protein. It also remains a distinct possibility that the CUE domain is required for the interaction between FANCD2 and another ubiquitinated DNA damage response protein.
The importance of interactions between ubiquitin and UBD containing proteins in the cellular DNA damage response has become increasingly evident. For example, the interaction between UBZ domains on Y-family polymerases and monoubiquitinated PCNA is essential for the polymerase switch that facilitates TLS.24,35 An intramolecular interaction between monoubiquitinated lysines in the carboxy-terminus of pol η and a UBZ domain in its amino-terminus promotes a closed conformation that precludes its interaction with PCNA.24 On exposure to DNA damaging agents, pol η is deubiquitinated leaving the UBZ free to interact with monoubiquitinated PCNA. Recent reports have also demonstrated a direct role for UBDs in the FA-BRCA pathway. The FAN1 and FANCP/SLX4 proteins harbor UBZ domains that are essential for cellular resistance to ICLs. Importantly, recruitment of these nucleases to sites of DNA damage relies on an interaction between their UBZ domains and monoubiquitin covalently linked to FANCD2.16-18,36 Furthermore, the newly identified FAAP20 protein has a UBZ4 domain that mediates binding to monoubiquitinated REV1 to promote TLS.37-39 Thus, FANCD2 represents the fourth FA pathway protein to harbor a functional UBD. Although there are important similarities between FANCD2 and pol η, there are also intriguing differences in the ubiquitin-dependent regulation of these proteins. For example, DNA damage stimulates FANCD2 monoubiquitination while it results in an inhibition of pol η monoubiquitination.4,24 Furthermore, although FANCD2 is monoubiquitinated on a single lysine residue K561, multiple lysine residues of the PCNA-interaction region of pol η can be monoubiquitinated.4,24 Indeed, Bienko et al speculate that in a situation where monoubiquitination leads to an intramolecular association between a UBD and ubiquitin in cis, monoubiquitination may not be restricted to a single lysine but rather may occur on any lysine residue in the region.24 Conversely, for homo or heterodimerization processes facilitated by UBD-ubiquitin associations in trans, as is the case for FANCD2 and FANCI, monoubiquitination of specific K residues may be critical. Consistent with this hypothesis, FANCI has been demonstrated to both stimulate FANCD2 monoubiquitination and to restrict FANCD2 monoubiquitination to K561 in vitro.40
Taken together our results establish a novel role for noncovalent ubiquitin signaling in the regulation of the FANCD2 protein, and add new insight into the structure and function of a poorly understood protein with important implications for bone marrow failure and cancer susceptibility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Howlett and Sun laboratories for helpful discussions.
This work was supported by a Leukemia Research Foundation New Investigator grant (N.G.H.); Rhode Island IDeA Network of Biomedical Research Excellence (RI-INBRE) grant P20RR016457-09 from the National Center for Research Resources (Zahir A. Shaikh, PI/NGH, faculty development); National Science Foundation EPSCoR grant no. 1004057, and National Institutes of Health/National Heart, Lung, and Blood Institute grants R21HL095991 (N.G.H.) and R01HL101977 (N.G.H.).
National Institutes of Health
Authorship
Contribution: M.A.R. designed experiments, performed research, analyzed data, and wrote the paper; F.W.K., E.A.V., and M.M. performed research and analyzed data; and N.G.H. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Niall G. Howlett, Department of Cell and Molecular Biology, University of Rhode Island, 379 Center for Biotechnology and Life Sciences, 120 Flagg Rd, Kingston, RI 02881; e-mail: nhowlett@mail.uri.edu.