To the editor:
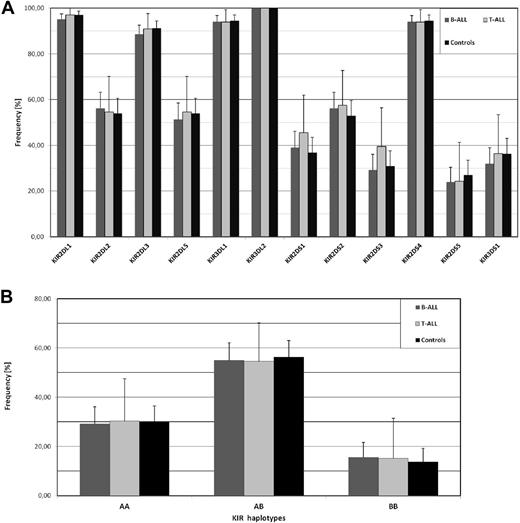
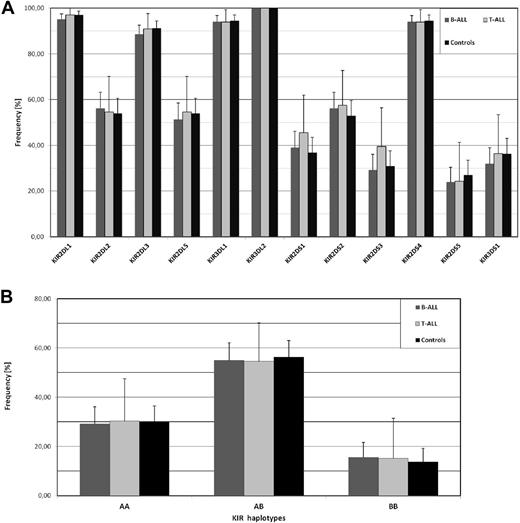
In a recent report, Almalte et al described novel associations between childhood acute lymphoblastic leukemia (ALL) and killer immunoglobulin-like receptor (KIR) genes in a case-control study including mostly French-Canadian patients.1 The study was limited to the analysis of stimulatory KIR (KIR-S) and impressively, all of the 6 different KIR-S exhibited a strongly reduced frequency in the patient cohort. We performed a similar analysis in a cohort of childhood B-ALL (n = 185) and T-ALL (n = 33) patients of European origin (92% German, recruitment 1992-2012) from the pediatric oncology center in Düsseldorf, but also included inhibitory KIR, which enabled the identification of extended KIR genotypes. As shown in Figure 1A, none of the KIR-S genes exhibited a significant frequency deviation from our ethnically matched control cohort. Our control group exhibited comparable KIR-S frequencies to the French-Canadian control group from Almalte et al1 except for KIR2DS5, which was unusually high in the Canadian study also when compared with other white cohorts from France, Germany, or the United Kingdom (data available at www.allelefrequencies.net). Because the strongest association in that study was seen for KIR2DS2, we looked for the frequency of the inhibitory KIR2DL2, which is in strong linkage disequilibrium with KIR2DS2. Again no decreased frequency of KIR2DL2 was found in our ALL cohort. The data from Almalte et al also implicate that the frequency of group A KIR haplotypes, which are abundant in white populations and harbor only a single KIR-S, would be much higher in ALL patients. Again our analysis does not show any significant difference between patients and controls (Figure 1B). Further analysis of telomeric and centromeric KIR haplotypes2 as well as the cumulative number of stimulatory KIR genes did not reveal any significant difference to the control cohort (data not shown).
No association of childhood ALL with KIR gene frequencies. (A) The frequency of 6 inhibitory KIR genes (KIR2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 3DL2) and 6 stimulatory KIR genes (KIR2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DS1) was analyzed in ALL patients. Our study population (0-18 years) consisted of 185 children with B-ALL (dark gray columns) and 33 children with T-ALL (light gray columns) of European origin. The ethnically matched control group (black columns) consisted of 204 unrelated randomly selected healthy volunteers. PCR-based KIR genotyping was performed as described by Vilches et al.3 As an additional quality control, 10% of samples were randomly selected and analysis repeated with an independent KIR typing protocol as described by Uhrberg et al.4 Similarly, all samples exhibiting rare KIR genotypes (frequency < 0.5%) were controlled in this way.4 Samples with discordant typing results (n = 9) were excluded from the analysis. (B) Distribution of group A and B KIR haplotypes according to previous definitions.5 Statistical significance was tested by 2-sided Student t test and 95% confidence intervals are indicated.
No association of childhood ALL with KIR gene frequencies. (A) The frequency of 6 inhibitory KIR genes (KIR2DL1, 2DL2, 2DL3, 2DL5, 3DL1, 3DL2) and 6 stimulatory KIR genes (KIR2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DS1) was analyzed in ALL patients. Our study population (0-18 years) consisted of 185 children with B-ALL (dark gray columns) and 33 children with T-ALL (light gray columns) of European origin. The ethnically matched control group (black columns) consisted of 204 unrelated randomly selected healthy volunteers. PCR-based KIR genotyping was performed as described by Vilches et al.3 As an additional quality control, 10% of samples were randomly selected and analysis repeated with an independent KIR typing protocol as described by Uhrberg et al.4 Similarly, all samples exhibiting rare KIR genotypes (frequency < 0.5%) were controlled in this way.4 Samples with discordant typing results (n = 9) were excluded from the analysis. (B) Distribution of group A and B KIR haplotypes according to previous definitions.5 Statistical significance was tested by 2-sided Student t test and 95% confidence intervals are indicated.
Given the technical challenges associated with PCR-based KIR genotyping, which is due to the strong similarity between KIR genes and the increasing number of alleles, it is generally helpful to assess extended KIR genotypes when performing case-control studies. Because of the strong linkage disequilibrium between several pairs of KIR, the knowledge of KIR genotypes provides an important plausibility control for KIR typing results. Moreover, in our experience historic patient sample collections can be particularly challenging for KIR typing, leading to decreased amplification efficiency compared with high-quality control samples. Given the consistently decreased frequencies of all KIR-S genes in the Almalte et al study,1 it would be highly desirable to know inhibitory KIR gene frequencies in this cohort, which would help to understand how the distribution of KIR genotypes is affected. Unfortunately, PCR primers and amplification conditions used for KIR typing were not specified. The lack of these data in the study by Almalte et al makes it difficult to assess the observed discrepancies between the 2 studies.
In summary, we could not confirm the association of KIR-S genes with the risk of childhood ALL in our cohort and would generally recommend the assessment of extended KIR genotypes when performing case-control studies.
Authorship
Acknowledgments: The authors thank all parents who gave their consent to use the biologic material from their minors. They also thank Carlos Vilches (Inmunogenética–HLA, Hospital University Puerta de Hierro, Madrid, Spain) for helpful comments and advice.
This work was supported by the Deutsche Krebshilfe e.V. (to M.U., R.M. and A.B.).
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Markus Uhrberg, Institute for Transplantation Diagnostics and Cell Therapeutics, Heinrich Heine University, Moorenstr 5, Düsseldorf, Germany 40225; e-mail: uhrberg@itz.uni-duesseldorf.de.