Abstract
Chromatin accessibility plays a key role in regulating cell type specific gene expression during hematopoiesis but has also been suggested to be aberrantly regulated during leukemogenesis. To understand the leukemogenic chromatin signature, we analyzed acute promyelocytic leukemia, a subtype of leukemia characterized by the expression of RARα-fusion proteins, such as PML-RARα. We used nuclease accessibility sequencing in cell lines as well as patient blasts to identify accessible DNA elements and identified > 100 000 accessible regions in each case. Using ChIP-seq, we identified H2A.Z as a histone modification generally associated with these accessible regions, whereas unsupervised clustering analysis of other chromatin features, including DNA methylation, H2A.Zac, H3ac, H3K9me3, H3K27me3, and the regulatory factor p300, distinguished 6 distinct clusters of accessible sites, each with a characteristic functional makeup. Of these, PML-RARα binding was found specifically at accessible chromatin regions characterized by p300 binding and hypoacetylated histones. Identifying regions with a similar epigenetic make up in t(8;21) acute myeloid leukemia (AML) cells, another subtype of AMLs, revealed that these regions are occupied by the oncofusion protein AML1-ETO. Together, our results suggest that oncofusion proteins localize to accessible regions and that chromatin accessibility together with p300 binding and histone acetylation characterize AML1-ETO and PML-RARα binding sites.
Introduction
Cell specific gene expression is regulated through a vast suite of epigenetic marks, such as chromatin modifications and DNA accessibility.1 Recently, genome-wide histone modification patterns have been extensively investigated in different cell types and organisms to understand the global histone marking and its influence on biologic processes.2-5 In the context of hematopoietic development and leukemia, various groups set out to characterize the global histone modification landscape.5-7 Although thought to be equally important, less effort has been put in understanding the role of DNA accessibility.8 High accessibility resulting from poor nucleosome occupancy has been suggested as a hallmark of (most) functional elements in the genome,9-11 highlighting its functional importance and endorsing more in-depth study of the global accessibility landscape.
Hematopoiesis is a complex differentiation system and it is tightly regulated by the interplay between cis elements and trans factors, including transcription factors and chromatin-modifying enzymes. Knowing both the binding pattern of a particular transcription factor as well as understanding the underlying chromatin has the potential to lead to better comprehension on how these factors regulate the different hematopoietic lineages as well as how the histone and accessibility patterns are changed in the context of leukemia. Different acute myeloid leukemia (AML) associated oncoproteins, such as PML-RARα t(15;17) and AML1-ETO t(8;21), are known to recruit repressive complexes, influencing different histone modification patterns and thereby altering the chromatin landscape, underlining the importance of chromatin dynamics in the context of different types of leukemia.
To determine the accessibility landscape, a variety of techniques are available, including assays based on restriction enzymes and DNaseI.9 Coupling these techniques with state-of-the-art high throughput sequencing technologies has made it possible to obtain high resolution pictures of the overall chromatin architecture and its dynamics with respect to different cellular phenomena. Indeed, high throughput studies based on the use of enzymatic activities12-14 or chromatin sonication15,16 have shed light on chromatin organization of primary cells, such as CD4+ T and CD34+ cells, mouse model cell lines, and human pancreatic islets.
Acute promyelocytic leukemia (APL) is a subtype of AML in which the epigenetic landscape is perturbed by the expression of the oncofusion protein PML-RARα.17 The ability of PML-RARα to distort the normal functioning of different chromatin-modifying factors can lead to changes in the local chromatin environment and hence the underlying DNA accessibility. Recent global studies of APL cells have given a comprehensive analysis of genome-wide PML-RARα binding sites and the all-trans retinoic acid (ATRA) induced changes in histone and DNA methylation profiles.6,18 These studies showed that on degradation of PML-RARα dramatic changes in histone modification patterns occur. Although these changes are expected to correlate with the accessibility landscape of the cells, it is currently unknown whether the epigenetic changes drive or follow changes in genome-wide accessibility and to what extent these chromatin alterations can be found in other oncoprotein-expressing AMLs.
Nuclease accessibility coupled with high throughput sequencing (NA-seq) has recently been introduced as a novel tool for the detailed structural analysis of chromatin alterations during differentiation of primary human CD34+ cells toward myeloid differentiation.13 In this study, we have established genome-wide nuclease accessibility maps for APL and AML cells using NA-seq methodology, and the changes occurring during ATRA-induced differentiation. In addition, we have analyzed the patterns of different epigenetic modifications, such as H2A.Z, H2A.Zac, H3ac, H3K4me3, H3K4me1, H3K9me3, and RNAPll and the regulatory factor p300. Our analysis allowed the identification of specific accessibility clusters in APL, which represent different functional clusters, based on co-occurrence of these factors. We also found that PML-RARα binds specific response elements in open chromatin regions and that PML-RARα bound response elements show an increased accessibility on ATRA treatment. Examining t(8;21) cells for the presence of the chromatin signature identified at PML-RARα binding sites allowed the identification of AML1-ETO binding sites. Together, these results suggest that oncoproteins localize to accessible regions and that chromatin accessibility together with a set of specific chromatin modifications are characteristics of AML1-ETO and PML-RARα binding sites.
Methods
Cell culture and preparation of accessible libraries
NB4 and SKNO-1 cells were cultured in RPMI medium with FCS at 37°C. For differentiation, NB4 cells were treated 48 hours with 10μM ATRA. For NA-seq, DNA regions were isolated as described before.13 In brief, isolated nuclei were treated with Nlalll and Hpall enzymes, purified DNA was reduced in size using Sau3AI, and processed for sequencing. As a control, similar libraries of total genomic DNA were prepared (see also supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Illumina sequencing of accessible libraries
Libraries were ligated to specialized adapters A and B (supplemental Table 1) having the sticky ends corresponding to Nlalll or Hpall and Sau3Al, respectively. The 150- to 600-bp size-selected fragments were purified, and Nlalll or Hpall containing fragments were purified using a biotin-streptavidin capture.19 PCR-amplified accessible libraries were sequenced using the Illumina Solexa platform. Sequenced reads were mapped to the human genome HG18 using ELAND (see also supplemental Methods).
NA-seq data analysis
Accessible regions were defined by merging all uniquely mapped sequence reads within contiguous 300-bp windows from all sequencing runs of Nlalll-Sau3AI libraries. Tags were counted in these regions from each of the accessible libraries and genomic controls. Using the Fisher test,20,21 regions having more than a 2-fold difference between the accessible libraries over naked genomic DNA were called accessible. Correction for multiple testing was applied using the q-value method.22 For the Hpall libraries, the same analysis was performed. Biologic replicates were generated for all libraries, and the total number of accessible regions in proliferating NB4 cells was the added sum of all replicates. For comparison of NA-seq data of different AML patient blasts, we used the Fisher test with a fold change of 1.2 and a q-value of 0.5. Genomic annotation of the accessible regions and enrichment analysis were performed using the Genomatix (http://www.genomatix.de) tools from RegionMiner Annotation and Statistics and GenomeInspector, respectively. All NA-seq and ChIP-seq raw data files have been submitted to GEO (accession no. GSE30254); genomic locations of all accessible regions identified in this study are provided in supplemental Table 3.
CATCH clustering analysis
Epigenetic marks were clustered using the CATCH clustering algorithm23 (http://catch.cmbi.ru.nl). Clustering was performed using the highly accessible fraction of NA-seq identified regions in proliferating NB4 cells. CATCH was run with default settings and a resolution of 20 bp. Two major clusters were identified, of which one was further refined into subclusters (see also supplemental Methods).
Analysis of ATRA-treated NA-seq libraries
To define differential accessibility regions between ATRA-treated and nontreated NB4 cells, accessible regions were further merged within 700-bp windows (see also supplemental Methods). The Fisher test was used followed by correction for multiple testing (q-value method) and a fold change of > 2. The same threshold settings were applied both for the Nlalll and Hpall libraries.
Patient blasts
APL blasts were obtained from a patient with newly diagnosed AML having t(15;17). The sample consisted of > 80% bone marrow invasion and was a typical FAB M3. AML FAB M1 blasts were obtained from patients with newly diagnosed AML having no detectable translocation. These studies were approved by the SUN Ethical Committee (7028032003).
Browser visualization of NA-Seq libraries
F-seq24 was used for genome browser visualization of all NA-seq libraries.
ChIP
Results
Genome-wide mapping of the accessible regions in APL cells
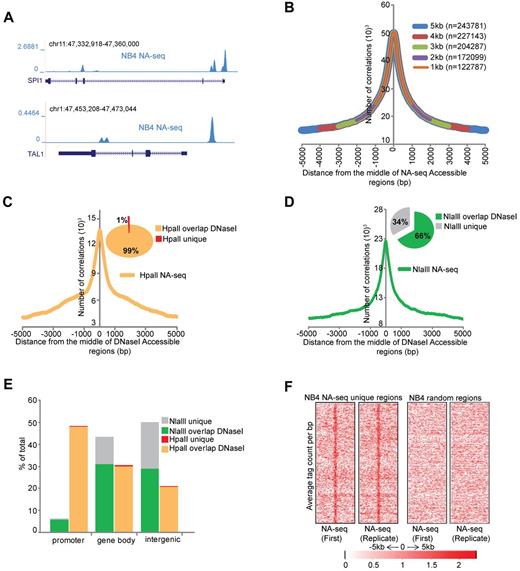
Chromatin accessibility is thought to be a main determinant in regulating gene expression during hematopoiesis, as it allows or prohibits the interaction of transcription-modulating factors with DNA. To map the nuclease accessible epigenome in APL, we used the nuclease accessible site sequencing (NA-seq) technique.13 For this, we generated restriction enzyme accessibility libraries through treatment of NB4 cell25 nuclei with the restriction enzymes Nlalll (CATĜ) or Hpall (ĈCGG), to provide significant coverage of the human genome.13 After this, the DNA fragments were further shortened in vitro by using a second enzyme, Sau3AI (GATC). The sticky end-containing fragments were ligated to linkers and sequenced on the Illumina platform (supplemental Figure 1A). After sequencing, tags were selected using the recognition motifs for the respective enzymes at the start of the sequence tag, thereby removing all nonspecific sequences. Biologic replicates were generated, and they showed for all datasets a very high Pearson correlation > 0.95 (supplemental Figure 1B), highlighting the reproducibility of the assay. As a control, naked genomic DNA was treated with the same enzymes and further processed for sequencing. To identify the accessible regions that were captured in our libraries, we merged all the uniquely mapped tags within 300 bp windows and defined accessible regions using the exact Fisher test,20,21 a multiple testing correction using the q-value method,22 and a threshold at a fold difference of 2. This analysis revealed 82 257 Nlalll and 24 912 Hpall marked nuclease accessible regions in proliferating NB4 cells (Table 1), for example, at key hematopoietic regulators, such as SPI1 and TAL1 (Figure 1A). Interestingly, most of these regions were enzyme specific, as only 4507 regions overlap between the NlaIII and HpaII sets (supplemental Figure 1C).
Identification of accessible regions in APL cells. (A) Overview of the accessible regions at the SPI1and TAL1 genes in NB4 cells identified by NA-seq using Nlalll and Hpall. (B) Enrichment of DNaseI hotspots within the NA-seq dataset. Enrichment was computed with a lowered threshold and different distances from the middle of the regions defined by NA-seq (see supplemental Methods). The number of overlapping regions for every size window are indicated. (C-D) Enrichment plots of NA-seq regions within a window of ± 5 kb of DNaseI-defined accessible regions in NB4 cells. Inset pie charts: The percentage of NA-seq regions that overlap with the DNAseI dataset. (C) HpaII data. (D) NlaIII data. (E) Genomic distribution of the unique and DNaseI hotspots overlapping NA-seq regions. Accessible sites were examined for their presence in promoter (−500 bp up to 100 bp downstream of transcription start site), gene body (intron and exon), or intergenic (everything else) regions. Analysis was performed using “Genomatix” tool for annotation and statistics, whereby 1.8% of the human genome consists of promoters, 44% gene bodies, and 54% intergenic regions. (F) Intensity plot showing tag enrichment from 2 biologic replicates within ± 5 kb windows from the center of the NA-seq regions that do not overlap with the DNaseI hotspots (left). The same number of random regions was plotted as control (right).
Identification of accessible regions in APL cells. (A) Overview of the accessible regions at the SPI1and TAL1 genes in NB4 cells identified by NA-seq using Nlalll and Hpall. (B) Enrichment of DNaseI hotspots within the NA-seq dataset. Enrichment was computed with a lowered threshold and different distances from the middle of the regions defined by NA-seq (see supplemental Methods). The number of overlapping regions for every size window are indicated. (C-D) Enrichment plots of NA-seq regions within a window of ± 5 kb of DNaseI-defined accessible regions in NB4 cells. Inset pie charts: The percentage of NA-seq regions that overlap with the DNAseI dataset. (C) HpaII data. (D) NlaIII data. (E) Genomic distribution of the unique and DNaseI hotspots overlapping NA-seq regions. Accessible sites were examined for their presence in promoter (−500 bp up to 100 bp downstream of transcription start site), gene body (intron and exon), or intergenic (everything else) regions. Analysis was performed using “Genomatix” tool for annotation and statistics, whereby 1.8% of the human genome consists of promoters, 44% gene bodies, and 54% intergenic regions. (F) Intensity plot showing tag enrichment from 2 biologic replicates within ± 5 kb windows from the center of the NA-seq regions that do not overlap with the DNaseI hotspots (left). The same number of random regions was plotted as control (right).
To validate the results, we compared the accessible sites detected in NB4 cells with publically available DNasel datasets from NB4 cells (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=189871639&c=chr21&g=wgEncodeUwDnase). For this, we scanned our dataset to examine whether a DNasel hotspot could be detected within 1- to 5-kb regions spanning the middle of the NA-seq accessible regions. We found a very significant enrichment of DNasel hotspots in our NA-seq dataset (Figure 1B), with 78% (243 781) of DNaseI-identified regions within a 5-kb distance of the NA-seq regions. Nearly all (99%) HpaII NA-seq regions overlapped with DNaseI hotspots, whereas only 66% of NlaIII regions overlapped (Figure 1C-D). These NA-seq “unique” regions (34% of Nlalll plus 1% of Hpall regions) were largely nonpromoter regions (Figure 1E). Although they do not overlap with the publically available DNaseI hotspots, we did not disqualify these regions because they showed a reproducible tag intensity among replicate experiments (Figure 1F) and because experimental system-specific parameters may impact the analysis resulting in apparent discrepancies between DNaseI and NA-seq, as previously noted between DNaseI and FAIRE.26
Examining the genomic distribution of the NA-seq regions revealed that 94% of Nlalll sites represent nonpromoter regions, whereas 6% are found within 500 bp upstream of transcription start sites (Figure 1E). Compared with the entire genome, of which 1.8% are promoter regions using this criterion, this still represents a 3-fold enrichment. By contrast, Hpall accessible regions are enriched 27-fold for promoters, in line with the expectation that the methylation sensitive restriction enzyme HpaII (CCGG) will preferentially cut within CpG islands of (most likely) active genes.
PML-RARα binds at accessible regions
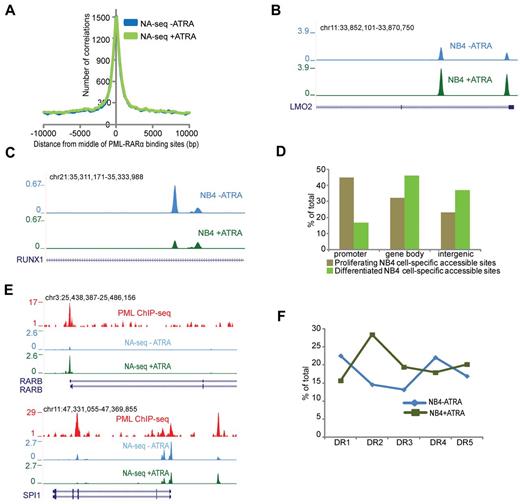
PML-RARα is the hallmark oncofusion protein associated with APL.27 ATRA treatment causes degradation of PML-RARα in NB4 cells, which leads to loss of proliferative capacity. Recently, the genome-wide binding pattern of PML-RARα was identified in NB4 cells.6,17 As PML-RARα has been suggested to induce a repressive chromatin environment,6,28,29 we wondered whether PML-RARα binding sites would compose inaccessible regions. Interestingly, we found that the majority of PML-RARα binding sites overlap with accessible regions in proliferating NB4 cells (Figure 2A), suggesting that the repressive capacity of PML-RARα is not translated toward chromatin inaccessibility. Because degradation of PML-RARα using ATRA triggers APL cell differentiation toward granulocytes,30 it allows to investigate whether the accessibility at PML-RARα binding sites is altered during this process. For this, we first treated NB4 cells with ATRA and identified 85 054 Nlalll and 27 752 Hpall accessible regions in these differentiated cells (Table 1). By applying the Fisher test and a stringent threshold for multiple correction (q = 1 × 10−06), we identified ∼ 2900 regions, which showed > 2-fold increased accessibility on ATRA-induced differentiation (eg, the LMO2 gene) and almost 2300 regions that showed reduction in accessibility (eg, the RUNX1 gene; Figure 2B-C). Genomic annotation of these dynamically accessible regions revealed that a substantial number of accessible promoters are specific for proliferating NB4 cells and that sites specific for differentiated NB4 cells can be mostly found in nonpromoter regions (Figure 2D).
Dynamic accessible regions and PML-RARα binding. (A) Enrichment of the accessible regions over the PML-RARα binding sites before (blue) and after (green) ATRA treatment. The plot represents the number of accessible regions enriched over a distance of ± 10 kb starting from the middle of the PML-RARα binding sites. (B) Overview of the LMO2 gene region in proliferating (blue) and ATRA-treated (green) NB4 cells. The depicted region shows an increase in accessibility on ATRA treatment. (C) Overview of the RUNX1 gene region in proliferating (blue) and ATRA treated (green) NB4 cells. The depicted region shows a decrease in accessibility on ATRA treatment. (D) Genomic annotation of ∼ 2300 and 2900 sites that display dynamic accessibility, respectively, gaining signal in proliferating and in ATRA-differentiated NB4 cells. Accessible sites were sorted in promoter (−500 bp up to 100 bp downstream of transcription start site), gene body (intron and exon), or intergenic (everything else) regions. Analysis was performed using the “Genomatix” tool for annotation and statistics. (E) Overview of NA-seq (Nlalll + Hpall) data at the PML-RARα targets RARβ and SPI1 showing accessibility changes on ATRA treatment. PML ChIP-seq indicating PML-RARα binding sites is plotted in red. NA-seq data in untreated cells are plotted in blue, whereas NA-seq data for ATRA treated cells are plotted in green. (F) DR motif analysis for PML-RARα binding sites, which show gain of accessibility after ATRA treatment (NB4 + ATRA), and for PML-RARα binding sites that lose accessibility (NB4 − ATRA).
Dynamic accessible regions and PML-RARα binding. (A) Enrichment of the accessible regions over the PML-RARα binding sites before (blue) and after (green) ATRA treatment. The plot represents the number of accessible regions enriched over a distance of ± 10 kb starting from the middle of the PML-RARα binding sites. (B) Overview of the LMO2 gene region in proliferating (blue) and ATRA-treated (green) NB4 cells. The depicted region shows an increase in accessibility on ATRA treatment. (C) Overview of the RUNX1 gene region in proliferating (blue) and ATRA treated (green) NB4 cells. The depicted region shows a decrease in accessibility on ATRA treatment. (D) Genomic annotation of ∼ 2300 and 2900 sites that display dynamic accessibility, respectively, gaining signal in proliferating and in ATRA-differentiated NB4 cells. Accessible sites were sorted in promoter (−500 bp up to 100 bp downstream of transcription start site), gene body (intron and exon), or intergenic (everything else) regions. Analysis was performed using the “Genomatix” tool for annotation and statistics. (E) Overview of NA-seq (Nlalll + Hpall) data at the PML-RARα targets RARβ and SPI1 showing accessibility changes on ATRA treatment. PML ChIP-seq indicating PML-RARα binding sites is plotted in red. NA-seq data in untreated cells are plotted in blue, whereas NA-seq data for ATRA treated cells are plotted in green. (F) DR motif analysis for PML-RARα binding sites, which show gain of accessibility after ATRA treatment (NB4 + ATRA), and for PML-RARα binding sites that lose accessibility (NB4 − ATRA).
To determine whether differentially expressed genes show differences in accessibility, we interrogated the promoter regions of previously identified up-regulated (1000) and down-regulated (900) genes in ATRA-treated NB4 cells6 and found enrichment of accessible regions at the center of all of these (supplemental Figure 2A-B). Generally, the promoter regions showed a stable accessibility status both in the proliferative as well as differentiated states, irrespective of expression dynamics of the corresponding genes (supplemental Figure 2A-B), except for a subset of approximately 100 promoters, for example, CCL2 and GFI1, which overlap with our dynamically changing accessible regions (supplemental Figure 2C-D). Together, these results suggest that changes in accessibility can influence gene expression but that other factors are involved in fine-tuning resulting gene expression.
Analysis of PML-RARα binding in relation to dynamically accessible regions revealed 134 PML-RARα binding regions, which had > 2-fold increased accessibility on ATRA-induced differentiation, whereas 213 PML-RARα regions showed the opposite pattern and overlapped with decreased accessibility regions. Regions that showed high increase in accessibility include well-known PML-RARα targets, such as the RARβ and SPI1 promoters (Figure 2E). Interestingly, for SPI1, we noticed decreased accessibility at the PML-RARα binding site in the third intron of the gene (Figure 2E), a region described as the transcription start site of an SPI1 negative regulating noncoding RNA.31
Because it is thought that on ATRA treatment PML-RARα, which shows promiscuous binding toward all variants of DR motifs AGGTCA(n)n,6 degrades and is replaced by the heterodimer RAR:RXR, which preferentially binds the DR2 (AGGTCA(n)2AGGTCA) and DR5 (AGGTCA(n)5AGGTCA) motifs, we investigated the presence of the DR response elements within the dynamic accessible regions. This analysis showed high enrichment for the retinoic acid response elements DR2 and DR5 in the accessible sites after ATRA treatment, whereas DR1 and DR4 repeats were enriched at accessible sites occupied by PML-RARα before ATRA-induced differentiation (Figure 2F).
Together, these results show that PML-RARα oncofusion binds to open chromatin regions and that genomic regions containing DR2 and DR5 are further opened on ATRA treatment, suggesting that at these positions PML-RARα is replaced by RAR:RXR.
Accessibility landscape in patient blasts and other leukemic subtypes
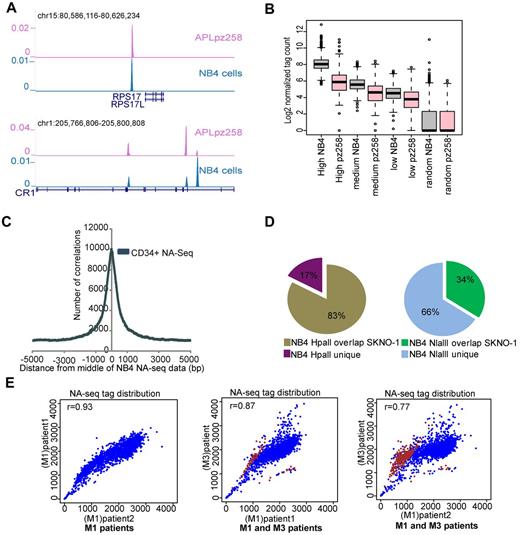
To further validate the NA-seq marked regions, we created a NA-seq library using the Nlalll enzyme in untreated APL patient blasts and found many similarities in the genomic location of accessible sites, for example, at the RPS17 and CR1 genomic loci (Figure 3A). We examined the APL tag density in the high, medium, and low accessible regions identified in proliferating NB4 cells (Table 2) and found a similar pattern of accessibility in terms of tag density in APL blasts as in NB4 cells (Figure 3B), whereas in comparison with a previously published NA-seq dataset from CD34+ cells,13 we found an overlap with 60 178 (∼ 54%) of our accessible regions identified in NB4 cells (Figure 3C), indicating differences in accessible regions between the APL and CD34+ cell types.
Differential accessibility between leukemic subtypes. (A) Overview of Nlalll-identified accessible regions at the RPS17 and CR1 genomic regions in APL (purple) as well as in NB4 cells (blue). (B) Correlation of accessible regions (high, medium, and low) and a set of randomly chosen regions in proliferating NB4 cells with the accessibility library in an APL patient sample. Box plot showing the normalized tag count for NB4 proliferating cells and for an APL patient sample (pz258) in accessible regions as defined in NB4 cells. (C) Enrichment of accessible regions defined by NA-seq in CD34+ cells within NA-seq defined NB4 accessible regions. Enrichment was investigated from the middle of the NB4 NA-seq dataset to a distance of ± 5 kb. (D) Overlap of accessible regions defined by NA-seq in APL (NB4 cells) and t(8;21) AML (SKNO-1 cells). Left panel: HpaII data. Right panel: NlaIII data. (E) Correlation of accessibility between different AML patient samples. In scatter plots, the NA-seq tag count was plotted in windows of 1.2 Mb over the whole genome (blue). Differential accessible regions between M1 and M3 patients and bound by PML-RARα are highlighted (brown). The correlation coefficient between pairs of datasets is depicted in the top left corner of each scatter plot.
Differential accessibility between leukemic subtypes. (A) Overview of Nlalll-identified accessible regions at the RPS17 and CR1 genomic regions in APL (purple) as well as in NB4 cells (blue). (B) Correlation of accessible regions (high, medium, and low) and a set of randomly chosen regions in proliferating NB4 cells with the accessibility library in an APL patient sample. Box plot showing the normalized tag count for NB4 proliferating cells and for an APL patient sample (pz258) in accessible regions as defined in NB4 cells. (C) Enrichment of accessible regions defined by NA-seq in CD34+ cells within NA-seq defined NB4 accessible regions. Enrichment was investigated from the middle of the NB4 NA-seq dataset to a distance of ± 5 kb. (D) Overlap of accessible regions defined by NA-seq in APL (NB4 cells) and t(8;21) AML (SKNO-1 cells). Left panel: HpaII data. Right panel: NlaIII data. (E) Correlation of accessibility between different AML patient samples. In scatter plots, the NA-seq tag count was plotted in windows of 1.2 Mb over the whole genome (blue). Differential accessible regions between M1 and M3 patients and bound by PML-RARα are highlighted (brown). The correlation coefficient between pairs of datasets is depicted in the top left corner of each scatter plot.
To assess the variability in accessibility between different AMLs, we also generated an accessibility profile in SKNO-1 cells, which represent an AML M2 subtype and harbor a t(8,21) translocation. Comparison of NB4 and SKNO-1 accessible regions showed a very high overlap of 83% at Hpall marked accessible promoters between the 2 cell types (Figure 3D). This significant overlap was also observed for the NlaIII marked promoters (supplemental Figure 3) whereas for the entire NlaIII set, composing mostly nonpromoter regions, an overlap of only 34% was found (Figure 3D). This difference in accessibility architecture at Nlalll-marked nonpromoter elements compared with HpaII-marked promoter elements suggests that especially the nonpromoter elements compose the crucial functional regions that regulate cell type identity.
To further analyze the heterogeneity between different AML patients, we generated genome-wide NlaIII accessibility profiles of 2 AML M1 patient cells. As the limited amount of material precluded sequencing of genomic DNA for these AML patients, and hence the Fisher analysis used for NB4, we plotted the accessibility tag distribution over 1.2 Mb windows13 in the whole genome from all 3 patient samples (2 AMLs M1, 1 APL M3). We noticed a very high correlation between the tag distribution in the 2 AML M1 patients (r = 0.93), whereas the r value is lower when we compare the tag distribution between M1 and M3 patient blasts (Figure 3E), again suggesting cell type specificity in accessible sites.
To identify the regions that show differences in accessibility pattern between the M1 and M3 AML subtypes, we used the Fisher test (see “NA-seq data analysis”). Examining the regions that showed differential accessibility patterns between APL and AML M1 subtypes revealed a high overlap (> 74%) with our previously published6 genome-wide PML-RARα binding sites, specifically for the regions that show increased accessibility in APL patients compared with the AML M1 blasts (Figure 3E), suggesting that the difference in accessibility pattern between APL and other AML subtypes might to a large extend be connected to expression of the PML-RARα oncofusion protein.
Epigenetic landscape over regulatory regions in APL cells
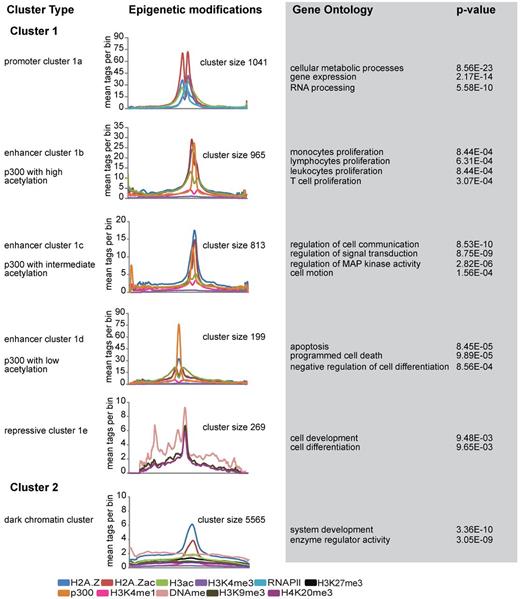
Our genomic distribution analysis revealed that the majority of Hpall-defined accessible sites are located in promoters. Indeed, analyzing the chromatin structure of HpaII sites revealed all previously reported features of promoter regions2,12,32 (supplemental Figure 4A-E; supplemental Methods). Our analysis also revealed that most differences in accessibility between cell types lie in nonpromoter regions (Figure 3D). To examine whether these nonpromoter regions compose different regulatory elements, we proceeded to discriminate subclasses of accessible nonpromoter regions based on their distinct epigenetic marking. To identify different epigenetic signatures, we used a clustering program called CATCH.23 As input for CATCH, we used the highly accessible NlaIII regions (Fisher test, P < .0001; Table 2) and a wide range of datasets on chromatin modification, including H2A.Z, H2A.Zac, H3ac, H3K4me3, H3K4me1, H3K9me3, DNA methylation, H3K27me3, H4K20me3, and RNAPll. In addition, p300 was included as a marker to locate enhancer elements.
CATCH clustering allowed the identification of 2 distinct clusters (Figure 4). Cluster 1 showed significant enrichment for a large variety of epigenetic marks, whereas cluster 2, containing > 5000 highly accessible regions, was only marked by H2A.Z and H2A.Z acetylation (Figure 4; supplemental Figure 5A-F). Cluster 1 was further divided into subclusters based on different characteristic epigenetic patterns. Among these, cluster 1a was characterized by increased H3K4me3 levels as well as increased levels of H2A.Z, H2A.Zac, H3ac, and RNAPII (Figure 4; supplemental Figure 5A). The high level of H3K4me3 suggested that this cluster represented the promoter fraction; and indeed, genomic annotation of these regions revealed their presence at promoter regions (data not shown). Independent analysis of all 5270 Nlalll marked promoters confirmed this conclusion (supplemental Figure 5G), highlighting the robustness of the clustering method. In contrast to cluster 1a, genomic annotation of the other 4 clusters (1b, 1c, 1d, and 1e) revealed enrichments toward nonpromoter regions, suggesting that these might represent enhancer elements. Interestingly, among these, 3 clusters were characterized by p300 enrichment. As several studies have identified p300 as an enhancer mark,33-35 we decided to name these 3 clusters enhancer clusters, which was further suggested by the significant presence of H3K4me1.35 The differential presence of histone modifications, in particular histone acetylation, allowed us to further define these 3 enhancer clusters. Enhancer cluster 1b is marked by high levels of H2A.Z and H3 acetylation, whereas enhancer cluster 1c showed decreased levels of both H3 and H2A.Z acetylation. The third enhancer cluster (1d) is characterized by high levels of p300 and low histone acetylation levels (Figure 4; supplemental Figure 5B-D). Functional characterization of the associated genes revealed that enhancer cluster 1b is highly enriched for genes involved in proliferation, whereas enhancer cluster 1c is characterized by genes involved in signal transduction and communication. In contrast, the enhancer cluster 1d showed high enrichment for genes related to apoptosis and regulators of cellular differentiation and contained genes, such as RUNX1 (supplemental Figure 5D) and GFI1. The last cluster (1e) was characterized by repressive chromatin marking, including DNA methylation, H3K9me3, and H4K20me3 (supplemental Figure 5E). Interestingly, analysis of the RNAPll occupancy in all CATCH-defined clusters revealed differential enrichments of RNAPll over different clusters. Apart from the promoters, enhancer cluster 1b and 1d were also enriched for RNAPll, whereas enhancer cluster 1c showed less RNAPll marking, comparable to repressive cluster 1e and cluster 2 (supplemental Figure 5H). Finally, cluster 2 was distinct in that it did not show any significant enrichment for histone marks, except H2A.Z and H2A.Z acetylation (Figure 4; supplemental Figure 5F). Although these results point toward H2A.Z as a general mark of chromatin accessibility, the exact role of the cluster 2 regions remains unclear.
Unsupervised clustering of highly accessible regions. Catch clustering analysis of highly accessible regions in proliferating NB4 cells based on profiles of H2A.Z, H2A.Zac, H3ac, p300, H3K4me3, RNAPII, H3K4me1, H3K27me3, H3K9me3, H4K20me3, and DNA methylation. The plots represent the average number of tag counts in 20-bp bins over 5-kb windows for each epigenetic mark in all clusters. Only the relevant epigenetic marks are shown in each cluster profile. Gene Ontology (GO) and P value were calculated using the online web tool “DAVID”:Database for Annotation, Visualization, and Integrated Discovery.
Unsupervised clustering of highly accessible regions. Catch clustering analysis of highly accessible regions in proliferating NB4 cells based on profiles of H2A.Z, H2A.Zac, H3ac, p300, H3K4me3, RNAPII, H3K4me1, H3K27me3, H3K9me3, H4K20me3, and DNA methylation. The plots represent the average number of tag counts in 20-bp bins over 5-kb windows for each epigenetic mark in all clusters. Only the relevant epigenetic marks are shown in each cluster profile. Gene Ontology (GO) and P value were calculated using the online web tool “DAVID”:Database for Annotation, Visualization, and Integrated Discovery.
Oncofusion proteins create a hypoacetylated chromatin environment at accessible p300 binding sites
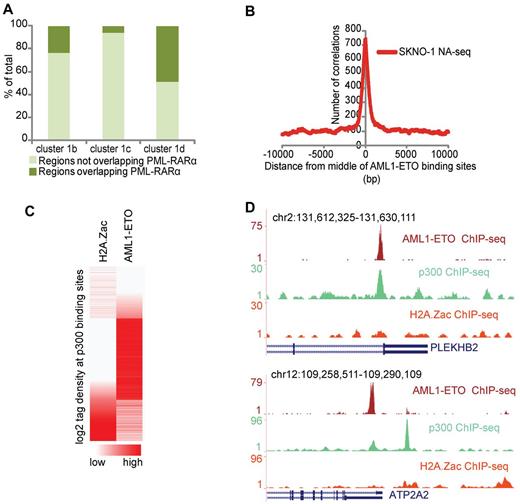
Aberrant recruitment of the epigenetic machinery and disruption of the normal histone modification patterns have been suggested as central activities involved in AML-associated oncofusion protein-mediated leukemogenesis. Our clustering analysis of regulatory regions revealed highest enrichment of PML-RARα binding sites in enhancer cluster 1d, which is characterized by high levels of p300 and low levels of acetylation (Figure 5A). These findings are concordant with previous results that showed that PML-RARα induces a hypoacetylated chromatin state.6 To investigate whether other oncofusion proteins, such as AML1-ETO, harbor similar chromatin characteristics, we overlapped a previously identified set of high confidence AML1-ETO binding sites37 and the NA-seq accessibility data in SKNO-1 cells. We found a very high enrichment of the accessible regions at AML1-ETO binding sites (Figure 5B), further confirming that oncofusion proteins, such as PML-RARα and AML1-ETO, bind preferentially to accessible regions.
PML-RARα and AML1-ETO binding sites share similar chromatin characteristics. (A) Percentage of PML-RARα binding sites in the different CATCH-defined enhancer clusters. Clusters 1b, 1c, and 1d from Figure 4 were examined for their relative PML-RARα presence. (B) Enrichment of the accessible regions identified in SKNO-1 cells at AML1-ETO binding sites. The plot represents the number of accessible regions enriched over a distance of ± 10 kb starting from the middle of the AML1-ETO binding sites. (C) H2A.Z acetylation and AML1-ETO tag density at p300 binding sites in SKNO-1 cells. p300 binding sites were called using MACS. (D) Overview of AML1-ETO (red), p300 (green), and H2A.Z acetylation (orange) at the PLEKHB2 and ATP2A2 genomic regions showing p300 and AML1-ETO binding but H2A.Z hypoacetylation.
PML-RARα and AML1-ETO binding sites share similar chromatin characteristics. (A) Percentage of PML-RARα binding sites in the different CATCH-defined enhancer clusters. Clusters 1b, 1c, and 1d from Figure 4 were examined for their relative PML-RARα presence. (B) Enrichment of the accessible regions identified in SKNO-1 cells at AML1-ETO binding sites. The plot represents the number of accessible regions enriched over a distance of ± 10 kb starting from the middle of the AML1-ETO binding sites. (C) H2A.Z acetylation and AML1-ETO tag density at p300 binding sites in SKNO-1 cells. p300 binding sites were called using MACS. (D) Overview of AML1-ETO (red), p300 (green), and H2A.Z acetylation (orange) at the PLEKHB2 and ATP2A2 genomic regions showing p300 and AML1-ETO binding but H2A.Z hypoacetylation.
To examine whether AML1-ETO binding sites have a similar chromatin signature as PML-RARα binding sites and whether such a signature can be used to predict AML1-ETO binding sites, we performed genome-wide ChIP-seq in SKNO-1 cells for p300 and H2A.Z acetylation, the 2 characteristic chromatin features of the accessibility cluster 1d identified in NB4 cells. We used MACS38 to call all p300 binding sites in SKNO-1 cells and analyzed the H2A.Z acetylation status. Correlating the acetylation status with AML1-ETO binding revealed that generally accessible sites with p300 and high H2A.Z acetylation do not have AML1-ETO binding, whereas accessible sites with p300 and low/intermediate levels of acetylation do have AML1-ETO binding (Figure 5C-D). Together, these results reveal that oncofusion proteins, such as AML1-ETO and PML-RARα, have similar chromatin characteristics and that these chromatin characteristics can be used to predict oncofusion protein binding sites.
Discussion
Chromatin structure and the accessibility of the underlying DNA are important determinants of proper execution of the regulation of gene expression programs in a given cell type.39,40 Different cis-regulatory elements having a crucial role in regulating transcriptional events are known to give access to their DNA sequence by having a more open chromatin conformation. This suggests that accessible regions of chromatin provide a basic scaffold for setting up a particular epigenomic environment.
In this study, we used NA-seq and identified > 100 000 accessible chromatin regions in proliferating APL cells. Analysis of the accessible sites showed that the use of only 2 different restriction enzymes already gives significant insight into accessible regions, encompassing promoters, gene bodies, and intergenic regions. HpaII preferentially cuts promoters of (most likely) active genes as indicated by the fact that 87% of them were RNAPII bound. In contrast, Nlalll cuts mostly intergenic regions, allowing us to look at putative enhancer regions.
DNaseI-seq and NA-seq agree for 99% of the promoter sites, which are highly accessible and generally regarded as open/nucleosome free. In contrast, for accessible sites located outside promoters, we observe only 66% concordance. The apparent discrepancies between DNaseI and NA-seq may reflect methodology specific variability.13,26 DNaseI-seq has the (dis)advantage that at low concentration or short digestion times, highly hypersensitive sites are easily digested; whereas at higher concentration or longer digestion times, the entire genome would be cut. Hence, a suitable window is determined that, given the nature of the kinetics of digestion, is inevitably a compromise. In contrast, restriction enzyme accessibility is sequence restricted but over digestion is not an issue. As a result, a lower extent of overlap is probably the result of the DNaseI dataset bias toward very accessible sites as it is kinetically impossible to simultaneously catch very accessible sites and less accessible sites with DNaseI. The available DNaseI datasets thus appear to be more biased on the hypersensitive promoters rather than on the nonpromoter sites that tend to be less accessible, also in our restriction endonuclease accessibility datasets, which may have hampered their confident identification by both methods.
PML-RARα expression is the hallmark oncofusion implicated in APL pathogenesis, and it has been postulated that PML-RARα binding causes a repressed chromatin state at its binding site through recruitment of corepressor complexes.17 Interestingly, in this study, we found the majority of PML-RARα binding sites in accessible chromatin bearing some hallmarks of active chromatin, similar to what has been reported recently for the glucocorticoid receptor.14 Moreover, we found enhanced accessibility on ATRA-induced degradation of PML-RARα on sites enriched for classic retinoic acid response elements, such as DR2 and DR5, suggesting that at these locations PML-RARα is replaced by the RAR:RXR heterodimer and that it is this heterodimer that is involved in chromatin opening. ATRA treatment triggers the process of differentiation in NB4 cells,30 allowing the identification of accessibility changes when cells go from proliferating to a differentiated state. We identified ∼ 5000 regions, which showed significant differences in accessibility between proliferating and differentiated NB4 cells. Genomic annotation showed that on ATRA-induced differentiation relatively more promoters are closed while more nonpromoter regions become accessible, suggesting that nonpromoter functional elements are important drivers of ATRA-mediated APL cell differentiation, consistent with the “repressive” activities of PML-RARα at nonpromoter elements.6
Like previous studies,2,10,40,41 we found a positive correlation between gene expression and high promoter accessibility. However, we found less accessibility changes at promoters corresponding to the genes that show changes in expression as the cells go from a proliferating to a differentiated state. These results suggest that gene expression positively correlates with accessibility but that the majority of the genes that are highly expressed as well as those that are poised for activation have accessibility repertoires, which are rather stable and, therefore, that other parameters than accessibility per se are involved in fine-tuning their expression levels.
Clustering analysis of accessible sites revealed 5 functional subclusters. From these, one had the characteristic H3K4me3 “active” promoter mark and showed enrichment for RNAPll, indicating a high correlation with transcriptional activity. Moreover, in this cluster, we found high levels of H2A.Z, H2A.Zac, and H3ac over the entire region, except for a dip in the middle, which probably represents the nucleosome free region.2 Of the 4 remaining clusters, 3 were marked with p300, which is known to bind enhancers.36 We further defined these enhancer clusters on the basis of the presence of histone acetylation and found one cluster with high levels of acetylation, one with intermediate levels and one with low levels. Interestingly, cluster 1d with low acetylation and very high p300 binding was enriched for PML-RARα binding sites, suggesting that this cluster is actively deacetylated through recruitment of histone deacetylase (HDAC) activities by PML-RARα. The role of this cluster in leukemia maintenance is further suggested by functional examination of the associated genes. This is enriched for genes involved in apoptosis and differentiation, such as GFI1 and RUNX1, and influencing expression of these genes might be crucial for leukemia development.
Extending the analysis to AML1-ETO, another AML-associated oncofusion protein, revealed a high enrichment of accessible regions at AML1-ETO binding sites, a chromatin characteristic similar to PML-RARα. Moreover, we found a hypoacetylated chromatin architecture, and colocalization with p300 at AML1-ETO binding sites, another common feature for both PML-RARα and AML1-ETO. As recently the acetylation of AML1-ETO by p300 has been suggested as a key step in t(8;21) leukemogenesis,42 the colocalization with p300 might also indicate a potential role of acetyltransferase activity in regulating the PML-RARα complex.
Based on our analysis, p300 does not seem to fully counteract HDAC activity toward histones in the presence of these oncofusion proteins as these are generally hypoacetylated. This might be because of a functional difference in that p300 can still acetylate AML1-ETO while simultaneously HDACs may deacetylate histones but not AML1-ETO. Alternatively, p300 acetylation of AML1-ETO may be restricted to particular subsets of genes. The simultaneous presence of histone acetyltransferases and HDACs is in accordance with the emerging picture of a balanced interplay of the antagonistic activities of histone-modifying enzymes, such as histone acetyltransferases and HDACs or methyltransferases and demethylases.43,44 Indeed, in a recent study, the dynamic balance of histone acetylation levels at chromatin regions was shown to be maintained by the activity of histone acetyltransferases/HDACs as well as histone demethylase LSD1-containing complexes.45 Based on these observations, we propose PML-RARα and AML1-ETO oncofusion proteins as stabilizers of deacetylase activity at their target regions in leukemic cells. As a result, the dynamic balance of the acetylation/deacetylation activity is disrupted and an aberrant acetylation profile is observed in these AML cells.
Together, these findings underscore the role of accessible chromatin regions as important regulatory elements that are subject to intense regulation by trans-acting factors to define cell type identity. Their aberrant regulation in AML at the epigenetic level might provide an entry point to test drugs that inhibit chromatin-modifying enzymes or complexes46 that can lead to proper execution of the differentiation program and eradicate the leukemic cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Francesco Bertolini and Ines Martin Padura for the AML samples.
This work was supported by the European Union (LSHC-CT-2005-518417 Epitron), the Dutch Cancer Foundation (KWF KUN 2009-4527), the Higher Education Commission of Pakistan, and The Netherlands Organization for Scientific Research (Vidi 917.11.322, J.H.A.M.). L.A. was supported by Associazione Italiana per la Ricerca sul Cancro (MFAG 4625). S.M. was supported by Associazione Italiana per la Ricerca sul Cancro, Cariplo, European Community (Epitron), Ministry of Health and University and Research. G.F. is Associazione Italiana per la Ricerca sul Cancro/Italian Foundation for Cancer Research fellow.
Authorship
Contribution: S.S., J.H.A.M., and C.L. designed and analyzed the experiments; S.S. performed the experiments; L.R., G.F., M.R., L.A., and M.S. contributed to some of the experiments; K.-J.F., F.G.N., and M.H. contributed to the bioinformatic analysis; S.S., J.H.A.M., H.G.S., C.L., and S.M. wrote the manuscript; H.G.S. and J.H.A.M. supervised the project; and all authors contributed to the manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joost H. A. Martens, Molecular Biology, Route 274, M850.03.035, Geert Grooteplein 26, Nijmegen 6525 GA, The Netherlands; e-mail: j.martens@ncmls.ru.nl; and Hendrik G. Stunnenberg, Molecular Biology, Route 274,M850.03.031, Geert Grooteplein 26, Nijmegen 6525 GA, The Netherlands; e-mail: h.stunnenberg@ncmls.ru.nl.