Abstract
T-box transcription factors T-bet (Tbx21) and Eomesodermin (Eomes) are critical players in CD8+ cytotoxic T lymphocyte effector function and differentiation, but how the expression of these transcription factors is regulated remains poorly defined. Here we show that dominant T cells directed toward human CMV, expressing significantly higher levels of T-bet with graded loss of Eomes expression (T-bethiEomeshi/lo), are more efficient in recognizing endogenously processed peptide-major histocompatibility complexes (pMHC) compared with subdominant virus-specific T cells expressing lower levels of T-bet and high levels of Eomes (T-betintEomeshi). Paradoxically, the T-bethiEomeshi/lo dominant populations that efficiently recognized endogenous antigen demonstrated lower intrinsic avidity for pMHC, whereas T-betintEomeshi subdominant populations were characterized by higher pMHC avidity and less efficient recognition of virus-infected cells. Importantly, differential endogenous viral antigen recognition by CMV-specific CD8+ T cells also correlated with the differentiation status and expression of perforin, granzyme B and K. Furthermore, we demonstrate that the expression of T-bet correlates with clonal expansion, differentiation status, and expression of perforin, granzyme B and K in antigen-specific T cells. These findings illustrate how endogenous viral antigen presentation during persistent viral infection may influence the transcriptional program of virus-specific T cells and their functional profile in the peripheral blood of humans.
Introduction
CD8+ T cells play a crucial role in controlling intracellular pathogens and proliferating malignant cells.1,2 After primary antigen exposure, acquisition and maintenance of polyfunctional capacity by antigen-specific T cells are required for an effective immune response.3 These functions include cytokine production, cytolysis of target cells, and the ability to proliferate and expand in response to repeated exposure to the cognate peptide epitope bound to MHC class I molecules.4,5 The loss of polyfunctional capacity is frequently seen in chronic viral infections where “exhausted” virus-specific CD8+ T cells fail to respond to the antigen challenge and lose the ability to use homeostatic cytokines, such as IL-7 and IL-15, efficiently.6,7 There is a growing body of evidence which has shown that the T-box transcription factors Eomesodermin (Eomes) and T-bet act as “master regulators” of CD8+ T-cell immunity, including expression of various effector functions and memory T-cell differentiation.8–12 Animals with compound mutations in T-bet and Eomes display impaired ability to express cytolytic mediators (eg, perforin and granzymes), fail to lyse target cells efficiently, and are unable to control acute viral infections.9 In addition, T-box transcription factors have also been shown to play a crucial role in the development of autoimmune diseases, including type 1 diabetes and myocarditis.13,14 More recently, other transcription factors, such as Runx3, which regulate Eomes expression and effector T-cell function, including IFN-γ, perforin, and granzyme B have also been identified as critical players in CD8+ T cell-mediated immunity.15 These studies highlight the intricate complexity of transcriptional regulation of adaptive immunity and its potential importance in protective immunity.
Although over the past decade the biology of T-box and other transcriptional factors has been extensively studied in murine models, recent studies have emphasized the possible significance of these regulators in the context of human infection and diseases.16–18 Nevertheless, the precise mechanism by which expression of these transcriptional master regulators is controlled remains one of the major conundrums of CD8+ T-cell biology. In particular, it is not known whether the interaction of T-cell receptor with peptide-MHC (pMHC) complex through endogenous processing of cognate peptide epitopes plays any role in modulating the expression of these transcription factors. In the present study, we show that T cells specific for human cytomegalovirus directed toward different viral antigens express differential patterns of T-box transcriptional factors and this expression is linked to their ability to recognize endogenously processed pMHC complexes on virus-infected cells. Paradoxically, the high levels of T-bet expression in the immunodominant CMV-specific T-cell populations were inversely correlated with avidity for pMHC complexes. The differential expression of T-bet was also associated with an altered differentiation status and expression of perforin, granzyme B, and granzyme K. Collectively, these results illustrate how endogenous viral antigen presentation during persistent viral infection may modulate the transcriptional program of antigen-specific T cells and their functional profile.
Methods
Healthy volunteers
A panel of 20 healthy CMV-seropositive donors were recruited for this study. Each volunteer used in this study was asked to sign the consent form as outlined in the Queensland Institute of Medical Research human ethics committee in accordance with the Declaration of Helsinki.
Establishment and maintenance of cell lines
Bulk T-cell cultures specific for the CMV-encoded epitopes, ELRRKMMYM (ELR), ELKRKMIYM (ELK), VTEHDTLLY (VTE), NLVPMVATV (NLV), and IPSINVHHY (IPS) were generated after stimulation of PBMCs with 1 μg/mL of cognate peptide, as previously described.19 Bulk T-cell cultures were maintained in RPMI containing 10% FBS (growth medium) containing recombinant IL-2 and 30% T-cell growth factor. The human fibroblast cell lines used in this study, JuSt (HLA A1 A2 B8), F57 (HLA A2), F61 (HLA A1 B35), and F66 (HLA A1 B8), were maintained in growth medium. For antigen presentation experiments, fibroblasts were treated with IFN-γ for 24 hours and then infected overnight with CMV at a multiplicity of infection of 5:1. EBV-transformed lymphoblastoid cell lines (LCLs) were established from seropositive donors by exogenous virus transformation of peripheral B cells. For antigen presentation experiments, LCLs were infected overnight with recombinant Vaccinia-expressing the CMV proteins, IE-1 or pp65, and then fixed with 2% paraformaldehyde.
Intracellular T-bet and Eomes analysis
To assess the expression of T-bet and Eomes in CMV-specific T-cell populations, PBMCs from CMV-seropositive donors were incubated for 4-5 hours with CMV-encoded epitopes in the presence of brefeldin A. Cells were incubated with peridinin chlorophyll cyanin (PerCP)–Cy5.5–conjugated anti-CD8 and then fixed and permeabilized using the eBioscience foxp3 staining kit (eBioscience). Cells were then incubated with PE-conjugated anti–T-bet and AlexaFluor-647–conjugated anti-Eomes (eBioscience). Cell acquisition was performed using a FACSCanto II (BD Biosciences). Postacquisition analysis was performed using FlowJo Version 8.8.3 software (TreeStar).
pMHC multimer staining and T-cell phenotyping
MHC-peptide multimers supplied by ProImmune and Immundex were used to detect epitope-specific CD8+ T cells. PBMCs were incubated for 20 minutes at 4°C with allophycocyanin- or PE-labeled MHC class I multimers specific for the HLA A2-restricted epitope NLV (pp65), the B*3501-restricted epitope IPS (pp65), the HLA A1 restricted epitope VTE (pp50), and the HLA B8 restricted epitopes ELR or ELK (referred to as ELR/K; IE-1). Cells were then incubated for a further 30 minutes at 4°C with PerCP-Cy5.5–conjugated anti-CD8 (eBioscience), FITC-conjugated anti-CD57 (BD Biosciences), and allophycocyanin or V450-conjugated anti-CD27 (BD Biosciences). Cells were then acquired using a FACSCanto II with FACSDiva Version 6.1.3 software. Postacquisition analysis was performed using FlowJo Version 8.8.3 software.
Intracellular perforin and granzyme analysis
To assess the expression of perforin and granzyme (Grz) A, B, and K in CMV-specific T-cell populations, PBMCs from CMV-seropositive donors were incubated with CMV-specific MHC class I multimers and PerCP-Cy5.5–conjugated anti-CD8, then fixed and permeabilized using the eBioscience foxp3 staining kit. Cells were then incubated with PE-conjugated anti-perforin (eBioscience), AlexaFluor-700–conjugated anti–granzyme B (BD Biosciences) and FITC-conjugated anti-granzyme A (BD Biosciences) or FITC-conjugated anti–granzyme K (Santa Cruz Biotechnology). Cell acquisition was performed using a FACSCanto II. Postacquisition analysis was performed using FlowJo Version 8.8.3 software. For the analysis of perforin, granzymes, and transcription factors, PBMCs were stained with PerCP-Cy5.5–conjugated anti-CD8, fixed and permeabilized as outlined in this section, then costained intracellularly for the effector molecules and transcription factors as outlined in this section. For costaining with perforin and granzyme B, a FITC-conjugated anti–T-bet (BioLegend) was used.
Intracellular cytokine staining
For the analysis of peptide sensitivity and endogenous antigen recognition by CMV-specific T cells, PBMCs or peptide epitope stimulated bulk T-cell cultures were incubated for 4-6 hours in the presence of FITC-conjugated anti-CD107a and monensin (BD Biosciences) with 10-fold serial dilutions of CMV-encoded epitopes, or at a responder to stimulator ratio of 5:1 with IFN-γ–activated HLA-matched fibroblasts infected with the HCMV strain TB40E-Bac (a kind gift from Dr Barry Slobedman, Westmead Millennium Institute, Sydney, Australia) or with the Ad-CMVpoly vector that encodes a polyepitope of CD8+ T-cell epitopes from CMV.20 For the analysis of endogenous antigen recognition in recombinant Vaccinia-infected LCLs, T cells were incubated at a responder to stimulator ratio of 5:1 with a panel of paraformaldehyde-fixed HLA-matched lymphoblastoid cell lines infected with recombinant Vaccinia virus encoding the HCMV antigens pp65 or IE-1 in the presence of anti-CD107a and monensin. Cells were then incubated with PerCP-Cy5.5–conjugated anti-CD8 or with CMV-specific MHC class I multimers before anti-CD8 staining. Cells were then fixed and permeabilized using a BD Cytofix/Cytoperm kit and incubated with allophycocyanin or AF700-conjugated anti–IFN-γ (BD Biosciences). Cell acquisition was performed using a FACSCanto II. Postacquisition analysis was performed using FlowJo Version 8.8.3 software (BD Biosciences PharMingen).
T-cell proliferation
PBMCs from HLA A1 and HLA A2 CMV donors were labeled with CFSE and stimulated with 1 μg/mL of VTE and NLV peptide in the presence or absence of 10 U/mL of recombinant IL-2 and/or 10 ng/mL of recombinant IL-12. Cells were harvested on day 5, and the expression of T-bet and Eomes, or granzyme B and perforin was assessed in divided and undivided cells as outlined in “Intracellular perforin and granzyme analysis.”
Statistical analysis
All statistical analyses were performed using GraphPad Prism Version 5 software. A 1-way ANOVA test was used to determine the significance of differences observed between the mean and SD from experimental samples. Differences were considered to be statistically significant where P < .05. Correlation analysis was performed using a nonparametric Spearman test.
Results
Differential expression of the transcription factors T-bet and eomesodermin in CMV-specific T-cell populations
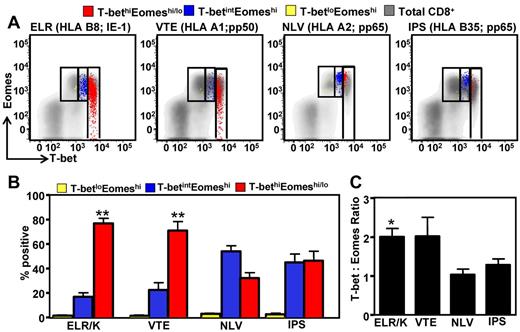
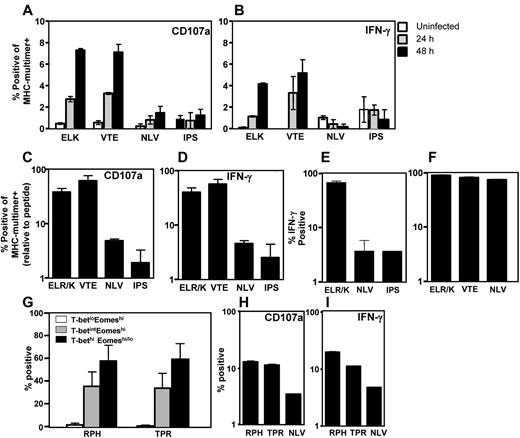
Recent observations, primarily in animal models, have demonstrated the critical role of the T-box transcription factors, T-bet and Eomes, in regulating effector function and the establishment of CD8+ T-cell memory.9,11,15 However, the precise mechanism underlying the regulation of expression of these transcription factors remains elusive. In the first set of experiments, we assessed the expression of T-bet and Eomes in CMV-specific T cells directed against 4 different viral epitopes ELR/K (IE-1), VTE (pp50), NLV (pp65), and IPS (pp65) restricted through HLA B8, A1, A2, and B35, respectively. PBMCs from a panel of 20 healthy virus carriers were stimulated with synthetic CMV peptide epitopes, and then CD8+ T cells and IFN-γ positive cells were assessed for the expression of T-bet and Eomes (Figure 1). CD8+ T cells displayed 3 distinct patterns of expression: T-betloEomeshi, T-betintEomeshi, and T-bethi.Eomeshi/lo (Figure 1A). T cells directed against the HLA B8-restricted ELR/K and HLA A1-restricted VTE epitopes had significantly higher levels of T-bet expression compared with the T cells specific for HLA A2-restricted NLV and HLA B35-restricted IPS epitopes (Figure 1A-B). Interestingly, a combined analysis of T-bet and Eomes revealed discordance in the expression of T-bet and Eomes in CMV-specific T cells (Figure 1B). Although very few CMV-specific T cells displayed a T-betloEomeshi phenotype, CD8+ T cells directed against ELR/K and VTE epitopes were predominantly T-bethi.Eomeshi/lo, and a significantly higher proportion of NLV- and IPS-specific T cells were T-betintEomeshi. Furthermore, analysis of the mean fluorescence intensity for T-bet and Eomes showed that the ratio of T-bet to Eomes expression in ELR/K- and VTE-specific populations was relatively higher compared with the NLV- and IPS-specific T cells (Figure 1C).
Differential expression of the transcription factors T-bet and Eomes in CMV-specific T cells. PBMCs from CMV-seropositive donors were incubated for 5 hours with cognate peptide epitopes, labeled with anti-CD8, and then assessed for intracellular expression of IFN-γ, T-bet, and Eomes. (A) Representative analysis of T-bet and Eomes expression in ELR/K-, VTE-, NLV-, and IPS-specific IFN-γ–producing cells is shown as an overlay of total CD8+ T cells. (B) Data represent the mean ± SEM of the proportion of CMV-specific T-cell populations displaying a T-betloEomeshi, T-betintEomeshi, or a T-bethi phenotype. Data represent analysis from at least 6 donors for each peptide tested. (C) Data represent the mean ± SEM of the ratio of mean fluorescent intensity of T-bet: mean fluorescent intensity of Eomes in different CMV-specific T-cell populations.
Differential expression of the transcription factors T-bet and Eomes in CMV-specific T cells. PBMCs from CMV-seropositive donors were incubated for 5 hours with cognate peptide epitopes, labeled with anti-CD8, and then assessed for intracellular expression of IFN-γ, T-bet, and Eomes. (A) Representative analysis of T-bet and Eomes expression in ELR/K-, VTE-, NLV-, and IPS-specific IFN-γ–producing cells is shown as an overlay of total CD8+ T cells. (B) Data represent the mean ± SEM of the proportion of CMV-specific T-cell populations displaying a T-betloEomeshi, T-betintEomeshi, or a T-bethi phenotype. Data represent analysis from at least 6 donors for each peptide tested. (C) Data represent the mean ± SEM of the ratio of mean fluorescent intensity of T-bet: mean fluorescent intensity of Eomes in different CMV-specific T-cell populations.
CMV-specific T cell expressing high levels of T-bet are more efficient in recognizing endogenously processed viral antigens
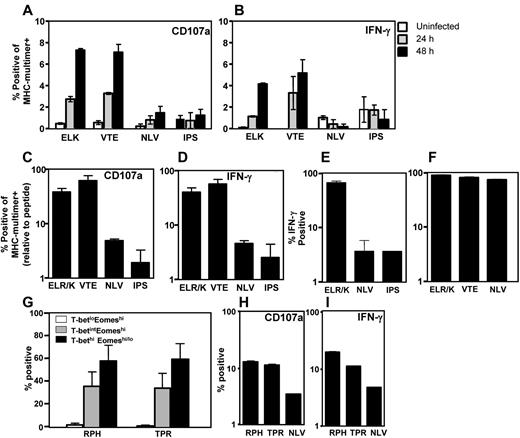
To delineate the possible mechanism for the distinct pattern of expression of T-box transcription factors in CMV-specific T cells, we next assessed the ability of these T cells to recognize endogenously processed epitopes from CMV-infected cells. PBMCs from healthy seropositive donors were incubated with CMV-infected (TB40E strain) HLA-matched human fibroblast cell lines and then assessed for degranulation (CD107a expression) and IFN-γ production. Data presented in Figure 2A-B show that ELR/K- and VTE-specific T cells displaying a T-bethi.Eomeshi/lo phenotype were more efficient in recognizing endogenously processed viral epitopes, whereas NLV- and IPS-specific T cells (T-betintEomeshi) poorly recognized virus-infected cells. ELR/K and VTE-specific T cells showed significant higher levels of CD107 mobilization (a surrogate marker for cytolytic function) and IFN-γ expression after exposure to CMV-infected target cells. These observations provided the first evidence on the potential link between endogenous antigen presentation and the expression of T-box transcription factors.
Differential recognition of endogenously processed CMV epitopes. (A-B) PBMCs from CMV-seropositive donors were incubated in the presence of monensin and anti-CD107a with a panel of TB40E-bac–infected fibroblast cell lines at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody and IFN-γ expression was assessed using an intracellular cytokine assay. Data represent the proportion of MHC-multimer+ cells expressing CD107a (A) or IFN-γ (B) after incubation with HLA-matched fibroblasts either uninfected or infected for 24 or 48 hours. (C-D) CMV-specific T cells were incubated in the presence of monensin and anti-CD107a with a panel of TB40E-bac–infected fibroblast cell lines at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody, and IFN-γ expression was assessed using an intracellular cytokine assay. Data represent the mean ± SEM from 3 independent experiments of the proportion of antigen-specific cells expressing CD107a (C) or IFN-γ (D) relative to peptide pulsed controls. (E) CMV-specific T cells were stimulated in the presence of monensin and anti-CD107a with a panel of HLA-matched paraformaldehyde-fixed LCLs infected with recombinant Vaccinia virus encoding either IE-1 or pp65 at a responder to stimulator ratio of 5:1. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody. Data represent the mean ± SEM of the proportion of antigen-specific cells generating IFN-γ. (F) CMV-specific T cells were incubated in the presence of monensin and anti-CD107a with the HLA A1 A2 B8-positive JuSt fibroblast cell lines infected overnight at a responder to stimulator ratio of 5:1 with AdCMVpoly. Data represent the mean ± SEM of the proportion of antigen-specific cells generating IFN-γ. (G) PBMCs from HLA B7 CMV-seropositive donors were incubated for 5 hours with the peptide epitopes RPHERNGFTVL (RPH) and TPRVTGGGAM (TPR), labeled with anti-CD8, and then assessed for intracellular expression of IFN-γ, T-bet, and Eomes. Data represent the mean ± SEM of the proportion of RPH or TPR-specific T cells displaying a T-betloEomeshi, T-betintEomeshi, or T-bethiEomeshi/lo phenotype. (H-I) RPH-, TPR-, and NLV-specific T cells were incubated in the presence of monensin and anti-CD107a with a TB40E-bac-infected HLA A2 B7-positive fibroblast cell line at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody, and IFN-γ expression was assessed using an intracellular cytokine assay.
Differential recognition of endogenously processed CMV epitopes. (A-B) PBMCs from CMV-seropositive donors were incubated in the presence of monensin and anti-CD107a with a panel of TB40E-bac–infected fibroblast cell lines at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody and IFN-γ expression was assessed using an intracellular cytokine assay. Data represent the proportion of MHC-multimer+ cells expressing CD107a (A) or IFN-γ (B) after incubation with HLA-matched fibroblasts either uninfected or infected for 24 or 48 hours. (C-D) CMV-specific T cells were incubated in the presence of monensin and anti-CD107a with a panel of TB40E-bac–infected fibroblast cell lines at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody, and IFN-γ expression was assessed using an intracellular cytokine assay. Data represent the mean ± SEM from 3 independent experiments of the proportion of antigen-specific cells expressing CD107a (C) or IFN-γ (D) relative to peptide pulsed controls. (E) CMV-specific T cells were stimulated in the presence of monensin and anti-CD107a with a panel of HLA-matched paraformaldehyde-fixed LCLs infected with recombinant Vaccinia virus encoding either IE-1 or pp65 at a responder to stimulator ratio of 5:1. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody. Data represent the mean ± SEM of the proportion of antigen-specific cells generating IFN-γ. (F) CMV-specific T cells were incubated in the presence of monensin and anti-CD107a with the HLA A1 A2 B8-positive JuSt fibroblast cell lines infected overnight at a responder to stimulator ratio of 5:1 with AdCMVpoly. Data represent the mean ± SEM of the proportion of antigen-specific cells generating IFN-γ. (G) PBMCs from HLA B7 CMV-seropositive donors were incubated for 5 hours with the peptide epitopes RPHERNGFTVL (RPH) and TPRVTGGGAM (TPR), labeled with anti-CD8, and then assessed for intracellular expression of IFN-γ, T-bet, and Eomes. Data represent the mean ± SEM of the proportion of RPH or TPR-specific T cells displaying a T-betloEomeshi, T-betintEomeshi, or T-bethiEomeshi/lo phenotype. (H-I) RPH-, TPR-, and NLV-specific T cells were incubated in the presence of monensin and anti-CD107a with a TB40E-bac-infected HLA A2 B7-positive fibroblast cell line at a responder to stimulator ratio of 5:1. Cells were then incubated with MHC-peptide multimers and anti-CD8 antibody, and IFN-γ expression was assessed using an intracellular cytokine assay.
It could be argued that differences in the recognition of virus-infected cells may be the result of the distinct activation status of CMV-specific T cells in vivo. To overcome this potential limitation, we generated a panel of ELR/K-, VTE-, NLV-, and IPS-specific T-cell lines after in vitro stimulation with cognate peptide. These effector cells were then incubated with a panel of HLA-matched fibroblasts infected with CMV. After incubation, MHC-multimer-specific populations were assessed for CD107a mobilization and IFN-γ production. As seen in the analysis of antigen recognition by T cells ex vivo, these experiments also revealed a contrasting pattern of T-cell recognition of virus-infected cells. CD8+ T cells specific for the ELR/K and VTE epitopes were most efficient in recognizing virus-infected cells (Figure 2C-D). In contrast, T cells specific for the epitopes IPS and NLV were comparably less efficient in recognizing virus-infected cells.
To further confirm that these differences were not associated with differential levels of antigen expression after CMV infection, a panel of HLA-matched EBV-transformed LCLs were infected with recombinant Vaccinia virus expressing either IE-1 (Vacc.IE1) or pp65 (Vacc.pp65) and then exposed to T cells specific for ELR/K, NLV, and IPS. Data presented in Figure 2E show that ELR/K-specific T cells were more efficient in recognizing Vaccinia-infected targets compared with both NLV- and IPS-specific T cells. To demonstrate that the differences in the efficiency of T-cell activation were attributable to the efficiency of epitope processing from the endogenous antigen, HLA A1 A2 B8-positive fibroblasts were infected with an adenoviral vector, AdCMVpoly, which encodes minimal preprocessed NLV, VTE, and ELR epitopes in a polyepitope sequence.20 After incubation with AdCMVpoly-infected fibroblasts, VTE, ELR/K, and NLV displayed a similar level of activation (Figure 2F), demonstrating that, in the context of the polyepitope, these epitope are processed and presented with similar efficiency. Finally, we assessed the functional programming and the efficiency of antigen presentation for 2 additional pp65 encoded epitopes. The pp65-encoded HLA B7 epitopes, RPHERNGVTL and TPRVTGGGAM, contain a high proportion of T-bethi Eomeshi/lo (Figure 2G) and were more efficiently presented compared with NLV (Figure 2H-I). Taken together, these analyses demonstrate that differences in the efficiency of endogenous presentation from full-length antigen correlate with differences in the transcriptional regulation of CMV-specific T cells.
CMV-specific T cells expressing higher levels of T-bet ex vivo display lower pMHC avidity
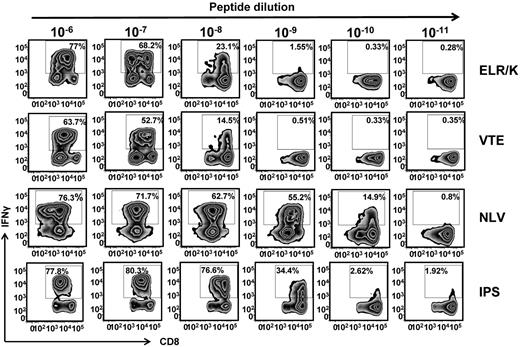
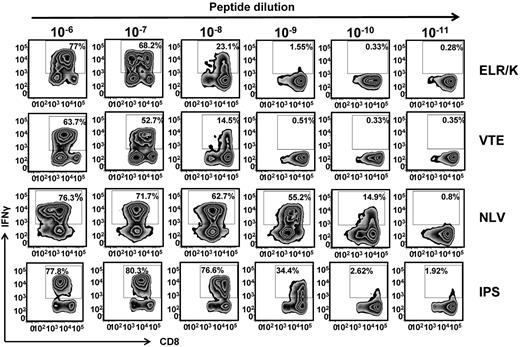
It has been established that T-cell avidity for antigen can shape the clonal dominance within an antigen specificity.21 To determine whether the recognition of endogenously processed epitopes was influenced by differential sensitivity to cognate peptide epitopes, CMV-specific T cells were incubated with serial dilutions of cognate peptide and assessed for IFN-γ. Interestingly, T cells specific for ELR/K and VTE, which expressed high levels of T-bet directly ex vivo and more efficiently recognized virus-infected cells were 10- to 100-fold less sensitive to cognate peptide compared with the NLV-specific and IPS-specific T cells (Figure 3). This difference in the avidity of T cells was not the result of the differential stability of the HLA-peptide complexes. Thermostability analyses of the HLA A1-VTE, HLA B8-ELR, and HLA A2-NLV complexes showed that these peptide epitopes bound to respective HLA molecules with comparable efficiency (data not shown).
Functional avidity of CMV-specific T cells. CMV-specific T cells were incubated for 5 hours with 10-fold serial dilution of cognate peptide epitopes. IFN-γ expression was then assessed using an intracellular cytokine assay. Representative flow cytometric analysis after stimulation of ELR/K, VTE, NLV, or IPS-specific T cells with serial dilutions of cognate peptide.
Functional avidity of CMV-specific T cells. CMV-specific T cells were incubated for 5 hours with 10-fold serial dilution of cognate peptide epitopes. IFN-γ expression was then assessed using an intracellular cytokine assay. Representative flow cytometric analysis after stimulation of ELR/K, VTE, NLV, or IPS-specific T cells with serial dilutions of cognate peptide.
Increased T-bet expression correlates with clonal expansion and a late-stage effector phenotype
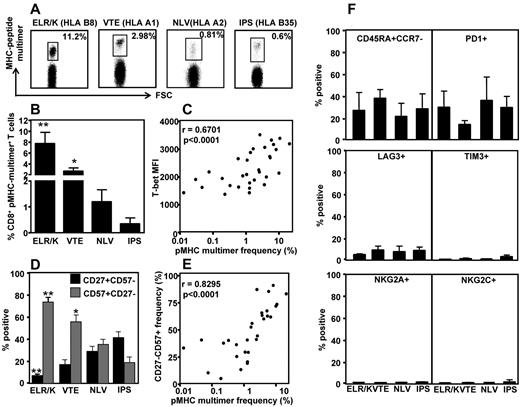
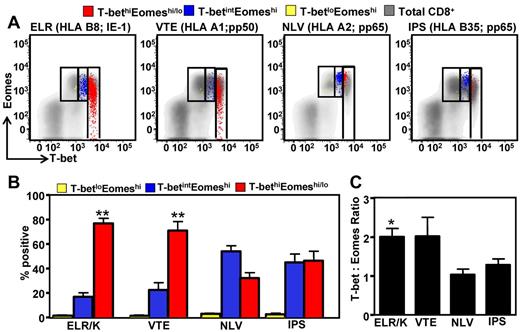
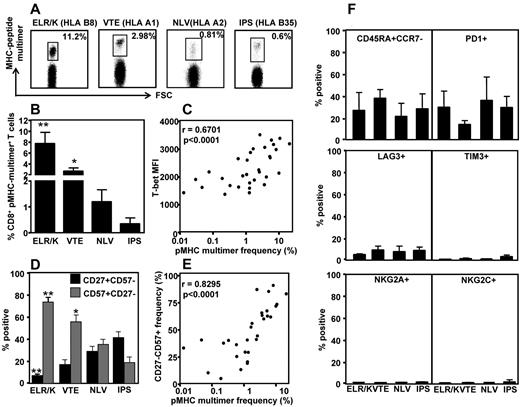
Previous studies have shown that persistent CMV infection in humans can induce large T-cell expansions that contribute to memory inflation.21 To investigate the association between T-cell frequency and transcriptional profile, the ex vivo frequency of ELR/K-, VTE-, NLV-, and IPS-specific T cells was assessed in PBMCs using pMHC multimers. A representative analysis for these epitopes is shown in Figure 4A. These analyses revealed that CMV-specific T cells displayed various degrees of clonal expansions (Figure 4B). Of particular note was the high frequency of T cells specific for the ELR/K epitope. The majority of HLA B8-positive donors generated ELR/K-specific responses in excess of 5% of total CD8+ T cells. In contrast, the frequency of T cells specific for the HLA B35-restricted IPS and HLA A2-restricted NLV epitopes was generally < 1% in the majority of the donors analyzed. Interestingly, pMHC multimer frequency showed strong correlation with the levels of T-bet expression in CMV-specific T cells (Figure 4C). We next assessed the expression of CD57, a marker for late-stage effector cells, and CD27, a marker expressed on less differentiated T cells on CMV-specific T cells (Figure 4D). These analyses revealed that the ELR/K- and VTE-specific T cells consistently displayed a predominantly late effector phenotype, characterized by a high proportion of CD27−CD57+ T cells and a low frequency of early memory CD27+CD57− cells (Figure 4D). Conversely, a higher proportion of the lower-frequency NLV- and IPS-specific T cells retained an early memory CD27+CD57− phenotype. More importantly, we noted a strong correlation between CD57 expression and pMHC multimer frequency (Figure 4E). The expression of other phenotypic markers, including CD45RA and CCR7, the NK cell receptors, NKG2A and NKG2C, and markers associated with chronic antigen stimulation, PD-1, TIM-3, and LAG3, did not correlate with epitope specificity or pMHC multimer frequency (Figure 4F). Taken together, these observations provide convincing evidence for a relationship between clonal T-cell expansion, effector T-cell differentiation, and the transcriptional regulation in virus-specific T cells.
Correlation between CMV epitope-specific T-cell frequency, T-bet expression, and CD27/CD57 expression by CMV-specific T cells. PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers, followed by fluorescently labeled anti-CD8-, CD27-, and CD57-specific antibodies. (A) Representation multimer analysis from 4 donors. (B) Data represent the mean ± SEM of pMHC multimer-specific CD8+ T cells in HLA-matched PBMCs. (C) Data represent the correlation between the frequency of pMHC multimer-specific cells and T-bet mean fluorescent intensity. (D) Data represent the mean ± SEM of epitope-specific T cells displaying either an early memory (CD27+CD57−) or late/terminal (CD27−CD57+) phenotype. (E) Data represent the correlation between the frequency of pMHC multimer-specific cells and the proportion of those cells expressing a late/terminal (CD8+CD27−CD57+) memory phenotype. (F) PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers, followed by fluorescently labeled anti-CD8, CD45RA, CCR7, PD1, LAG3, TIM3, NKG2A and NKG2C antibodies.
Correlation between CMV epitope-specific T-cell frequency, T-bet expression, and CD27/CD57 expression by CMV-specific T cells. PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers, followed by fluorescently labeled anti-CD8-, CD27-, and CD57-specific antibodies. (A) Representation multimer analysis from 4 donors. (B) Data represent the mean ± SEM of pMHC multimer-specific CD8+ T cells in HLA-matched PBMCs. (C) Data represent the correlation between the frequency of pMHC multimer-specific cells and T-bet mean fluorescent intensity. (D) Data represent the mean ± SEM of epitope-specific T cells displaying either an early memory (CD27+CD57−) or late/terminal (CD27−CD57+) phenotype. (E) Data represent the correlation between the frequency of pMHC multimer-specific cells and the proportion of those cells expressing a late/terminal (CD8+CD27−CD57+) memory phenotype. (F) PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers, followed by fluorescently labeled anti-CD8, CD45RA, CCR7, PD1, LAG3, TIM3, NKG2A and NKG2C antibodies.
Differential expression of T-bet correlates with the functional potential of CMV-specific CD8+ T cells
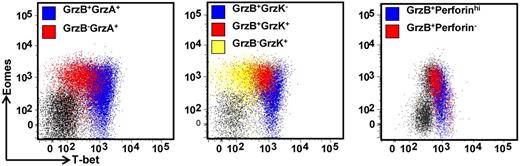
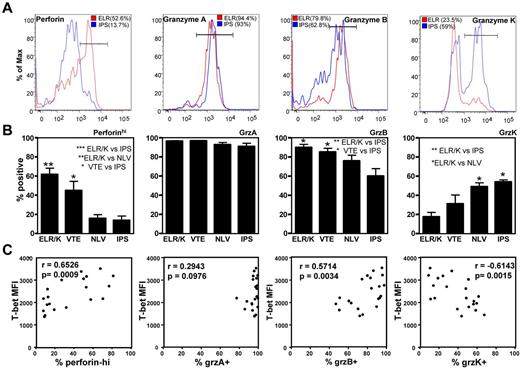
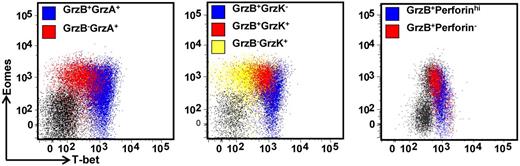
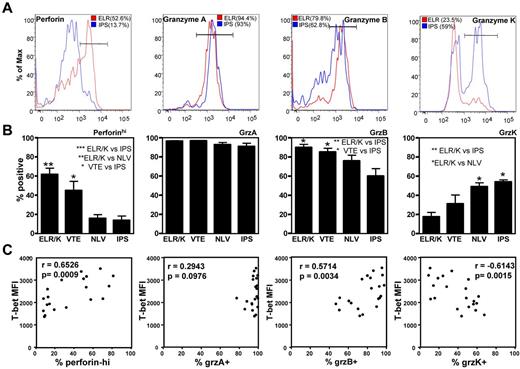
To further explore the potential impact T-box transcriptional factors have on T-cell function, we next assessed the expression of granzymes, A, B, and K and perforin in CD8+ T cells and CMV-specific CD8+ T cells. PBMCs were costained for the expression of T-bet and Eomes, and granzymes A, B, and K, and perforin. Granzyme or perforin-positive CD8+ T cells were then assessed for T-bet and Eomes expression. As is evident in Figure 5, there is a clear correlation between differential granzyme expression and the intensity of T-bet in CD8+ T cells. Although only granzymes A and K were detected in T-betloEomeshi cells, granzymes A, B, and K could be detected in T-betintEomeshi and only granzyme A and B expression in T-bethiEomeshi/lo cells. Similarly, high perforin expression was only detected in T-bethiEomeshi/lo cells. Analysis in CMV-specific populations also revealed significant differences in the expression of perforin, granzyme B, and granzyme K in CMV-specific T-cell populations directed against different epitopes (representative data showing analysis of perforin and granzyme expression in the ELR/K- and IPS-specific T cells from a single HLA B8 and B35-positive donor is shown in Figure 6A). T cells specific for ELR/K and VTE epitopes expressed high levels of perforin compared with NLV and IPS-specific T cells (Figure 6B). Although the majority of virus-specific T cells expressed both granzyme A and B, a significantly higher proportion of ELR/K and VTE-specific T cells were granzyme B-positive (Figure 6B). Conversely, a significantly greater proportion of NLV and IPS-specific T cells were positive for granzyme K (Figure 6B). Further analyses indicated that there was a strong correlation between T-bet MFI and the proportion of granzyme B+ and perforinhi CMV-specific T cells (Figure 6C). Interestingly, the percentage of granzyme A+ cells were almost identical for ELR/K, VTE, NLV, and IPS-specific T cells, and there was no correlation between the levels of T-bet expression and proportion of granzyme A+ T cells.
Differential expression of the effector molecules perforin and granzymes A, B, and K in CD8+ T cells correlates with the intensity of T-bet expression. PBMCs from a CMV-seropositive donor were stained with anti-CD8 and then assessed for the intracellular expression of T-bet, Eomes, granzymes A, B, and K, and perforin. Representative analysis of T-bet and Eomes expression in granzyme A, B, and K and perforin-producing cells is shown as an overlay of total CD8+ T cells.
Differential expression of the effector molecules perforin and granzymes A, B, and K in CD8+ T cells correlates with the intensity of T-bet expression. PBMCs from a CMV-seropositive donor were stained with anti-CD8 and then assessed for the intracellular expression of T-bet, Eomes, granzymes A, B, and K, and perforin. Representative analysis of T-bet and Eomes expression in granzyme A, B, and K and perforin-producing cells is shown as an overlay of total CD8+ T cells.
Differential expression of the effector molecules perforin and granzymes A, B, and K in CMV-specific T cells correlates with the intensity of T-bet expression. (A) PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers and anti-CD8 and then assessed for the intracellular expression of perforin and granzymes A, B, and K. (A) Histograms represent the analysis of perforin and granzyme A, B, and K expression in ELR/K and IPS-specific T-cell populations in a single donor. (B) Data represent the mean ± SEM of the proportion of perforinhi, granzyme A+, granzyme B+, or granzyme K+ epitope-specific T cells. (C) Data represent the correlation between the T-bet mean fluorescent intensity in CMV-specific T cells and the proportion of perforinhi, granzyme A+, granzyme B+, or granzyme K+ T cells.
Differential expression of the effector molecules perforin and granzymes A, B, and K in CMV-specific T cells correlates with the intensity of T-bet expression. (A) PBMCs from CMV-seropositive donors were stained with CMV-specific pMHC multimers and anti-CD8 and then assessed for the intracellular expression of perforin and granzymes A, B, and K. (A) Histograms represent the analysis of perforin and granzyme A, B, and K expression in ELR/K and IPS-specific T-cell populations in a single donor. (B) Data represent the mean ± SEM of the proportion of perforinhi, granzyme A+, granzyme B+, or granzyme K+ epitope-specific T cells. (C) Data represent the correlation between the T-bet mean fluorescent intensity in CMV-specific T cells and the proportion of perforinhi, granzyme A+, granzyme B+, or granzyme K+ T cells.
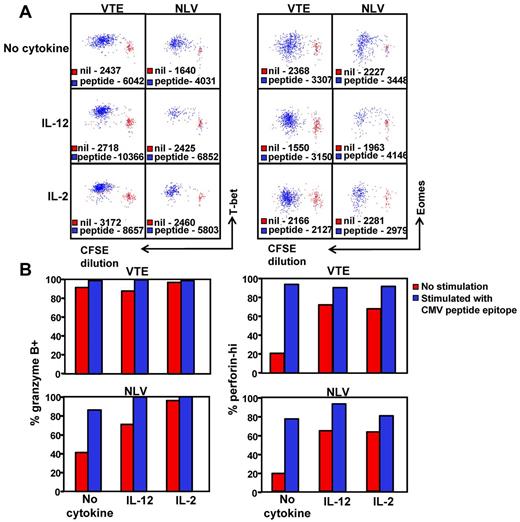
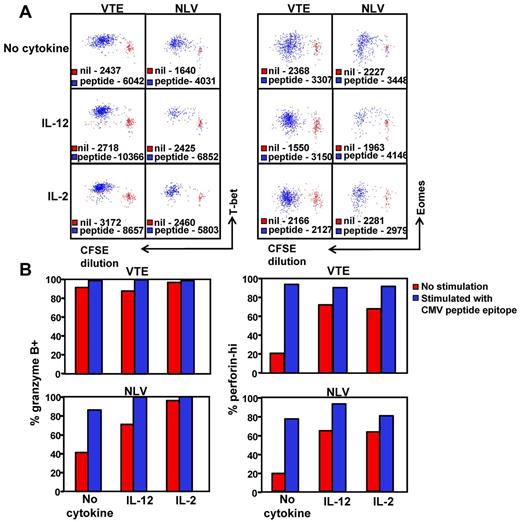
Given that high T-bet expression clearly correlates with a granzyme B+perforinhi phenotype, we next sought to determine whether the in vitro stimulation of T-betintEomeshi T cells would lead to an increase in T-bet expression and the transition to a granzyme B+perforinhi phenotype. PBMCs from an HLA A1 (VTE- T-bethiEomeshi) and an HLA A2 (NLV-T-betintEomeshi) positive donor were labeled with CFSE, then stimulated for 5 days with VTE and NLV, respectively, with and without either IL-2 and/or IL-12. NLV- and VTE-specific T-cell populations were then assessed for the expression of T-bet and Eomes (Figure 7A) or for granzyme B and perforin (Figure 7B). Stimulation with peptide led to an increase in the expression of both T-bet and Eomes in dividing NLV- and VTE-specific T cells (Figure 7A). This was concordant with an increase in the proportion of granzyme B+ and perforinhi T cells in both populations (Figure 7B). Although culture in the presence IL-2 or IL-12 led to an increase in the expression of T-bet, granzyme B, and perforin, peptide stimulation in the presence of IL-12 or IL-2 further enhanced the level of T-bet expression in both VTE and NLV-specific T cells, which correlated with a higher proportion of granzyme B+ perforinhi in NLV-specific T cells. Culture in the presence of either cytokine alone did not induce an up-regulation in the expression of Eomes and differentially affected the up-regulation of Eomes expression in the presence of peptide, particularly IL-2, which reduced the level of peptide induced Eomes up-regulation in both populations. These observations further emphasize the association between T-bet expression and the functional differentiation of CMV-specific T cells.
Induction of T-bet expression correlates with the transition to a granzyme B+perforinhi phenotype. (A) PBMCs from HLA A1 and HLA A2 donors were labeled with CFSE and then cultured with and without VTE or NLV for 5 days in the presence of IL-2, IL-12, or no cytokine. T-bet and Eomes expression in antigen-specific T cells was assessed as outlined in “Methods.” Data represent the mean fluorescence intensity of T-bet (left panel) or Eomes (right panel) expression in antigen-specific T cells stimulated with cognate peptide (blue) or incubated for 5 days without stimulation (red). (B) VTE- and NLV-specific T cells stimulated for 5 days with cognate peptide with and without cytokine were assessed for the expression of granzyme B and perforin. Data represent the proportion of granzyme B+ and perforinhi VTE and NLV-specific T cells, following in vitro culture with and without peptide.
Induction of T-bet expression correlates with the transition to a granzyme B+perforinhi phenotype. (A) PBMCs from HLA A1 and HLA A2 donors were labeled with CFSE and then cultured with and without VTE or NLV for 5 days in the presence of IL-2, IL-12, or no cytokine. T-bet and Eomes expression in antigen-specific T cells was assessed as outlined in “Methods.” Data represent the mean fluorescence intensity of T-bet (left panel) or Eomes (right panel) expression in antigen-specific T cells stimulated with cognate peptide (blue) or incubated for 5 days without stimulation (red). (B) VTE- and NLV-specific T cells stimulated for 5 days with cognate peptide with and without cytokine were assessed for the expression of granzyme B and perforin. Data represent the proportion of granzyme B+ and perforinhi VTE and NLV-specific T cells, following in vitro culture with and without peptide.
Discussion
The CD8+ T-cell repertoire generated after infection with latent/persistent viral infections, such as CMV, is directed toward a diverse array of antigens and often leads to accumulation of large effector memory T cells in humans that are unparalleled by other viral infections.22–24 T cells specific for certain epitopes are preferentially expanded and display various patterns of phenotypic and functional profiles. The precise mechanism for this differential response remains poorly defined. Nevertheless, a number of studies have proposed that selection forces, including pMHC-T-cell receptor avidity,21,24 precursor frequency recruitment and expansion,25 efficiency of antigen presentation,26,27 responsiveness to homeostatic cytokines,28,29 and structural constraints,30 have a significant impact on this process. More recently, the importance of T-box transcription factors (ie, T-bet and Eomes) in the differentiation and development of antiviral CD8+ T-cell responses have been recognized.9,11 Indeed, previous studies have shown that high T-bet expression is associated with long-term resilience, low expression of inhibitory receptors, and protection from CD8+ T-cell exhaustion.16,31 Data presented in this study extend these observations and demonstrate that the level of T-box transcription factor expression in virus-specific CD8+ T cells is associated with the efficiency of endogenous antigen presentation. Furthermore, expression of T-bet also correlates with the clonal expansion, phenotypic and functional profile of antigen-specific T cells.
How does endogenous antigen presentation regulate the expression of T-box transcription factors, which in turn impacts on the functional and phenotypic profile of antigen-specific T cells? CMV infection is a classic example of a persistent viral infection that induces accumulation of a population of virus-specific T cells, which display a phenotypic profile of activated cells, a phenomenon referred to as “memory inflation.”32 However, data presented here show that this memory inflation is highly discordant and efficient recognition of virus-infected cells was coincident with dominant expansion of T-bethiEomeshi/lo antigen-specific T cells, whereas T-cell specificities with lower precursor frequencies were less efficient in recognizing virus-infected cells and displayed a phenotype with T-betintEomeshi expression. Interestingly, these differentially expanded T-cell populations displayed opposing exogenous peptide avidity whereby T cells which more efficiently recognized endogenous antigen were consistently less sensitive to cognate peptide. Although the precise mechanism for the selection of T-bethiEomeshi/lo lower avidity dominant T cells is not known, it is plausible that, after priming, the increased antigen threshold required to maintain these effector cells would be met through more efficient endogenous presentation of viral epitopes. Conversely, it is apparent that the efficiency of antigen presentation places constraints on the T-cell repertoire specific for epitopes processed less efficiently, which by necessity have to select high-avidity clonotypes.
Previous studies have shown that inflammatory signals (eg, IL-12) not only enhance T-bet expression but might also repress Eomes and thus play a dominant role in regulating memory/effector T-cell potential.10,12 It is worth noting that murine CMV infection produces copious amounts of IL-12, which is primarily secreted by “newly” activated mature CD8α+ dendritic cells. Therefore, inflammatory stimuli, such as IL-12, probably play a significant role in controlling the expression of the T-box transcription factor after CMV infection.33,34 During the current investigation, the addition of IL-12 and, to a lesser extent, IL-2 further enhanced antigen driven up-regulation of T-bet in vitro, which is indicative of the role these cytokines play in the programming of human CD8+ T cells. Therefore, IL-12 production by dendritic cells probably plays a pivotal role in the up-regulation of T-bet expression in CMV-specific T cells, particularly during the priming phase. Nevertheless, the discordant pattern of expression of T-box transcription factors in CMV-specific T cells suggests that the interaction of T-cell receptor with pMHC complex on virus-infected cells will also influence the transcriptional programming of CD8+ T cells. However, it is currently unclear whether the discordant regulation of T-box transcription factors in different CMV-specific T cells is shaped during initial T-cell priming or after exposure to persistent antigen. During initial T-cell priming, cross-presentation by IL-12 secreting dendritic cells plays an important role in T-cell activation, and the abundance of antigen and the efficiency with which this antigen is cross-presented to CD8+ T cells will probably influence the initial transcriptional programming of responding naive T cells. Conversely recent studies have shown that, after the establishment of latency, the continuous stimulation of murine CMV-specific T cells by virus-infected nonhematopoietic cells is essential to ensure the maintenance of a functional effector CD8+ T-cell pool in the peripheral blood, which provides protection against viral reactivation.35,36 This persistent antigen exposure also probably contributes to the clonal expansion and acquisition of an effector phenotypic profile with strong expression of CD57, perforin, and granzyme B, which strongly correlate with T-bet expression. Thus, it is likely that a combination of persistent antigenic stimulation through endogenous antigen presentation and inflammatory signals regulate the expression of T-box transcription factors in antigen-specific T cells. These observations have important implications for designing immune-based strategies against persistent viral infections where a robust effector/memory T-cell response can be established through modulating endogenous antigen presentation and inflammatory signals.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NH&MRC), Queensland Cancer Fund, and the Australian Research Council. R.K. and S.R.B. were supported by NH&MRC (Senior Principal Research Fellowship and Senior Research Fellowship, respectively). J.R. was supported by the NH&MRC Australia (Fellowship). K.K.W. and S.-K.T. were supported by the Queensland Cancer Fund and Leukemia Foundation Australia (scholarships). D.E. and J.T. were supported by the NH&MRC (career development fellowships).
Authorship
Contribution: C.S. and R.K. designed the study and wrote the manuscript; C.S., D.E., S.G., K.K.W., V.D., J.T., S.-K.T., S.R. and Y.C.L. conducted various experimental studies; and J.R. and S.R.B. provided critical intellectual input and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rajiv Khanna, Queensland Institute of Medical Research, Tumour Immunology Laboratory, Department of Immunology, 300 Herston Road, Brisbane, Australia; e-mail: rajiv.khanna@qimr.edu.au.