Abstract
In the present study, we re-annotated von Willebrand factor (VWF), assigned its entire sequence to specific modules, and related these modules to structure using electron microscopy (EM). The D domains are assemblies of smaller modules visible as lobes in EM. Modules in the D-domain assemblies include von Willebrand D, 8-cysteine, trypsin inhibitor-like, E or fibronectin type 1-like domains, and a unique D4N module in D4. The D1-D2 prodomain shows 2 large connected assemblies, each containing smaller lobes. The previous B and C regions of VWF are re-annotated as 6 tandem von Willebrand C (VWC) and VWC-like domains. These 6 VWC domains correspond to 6 elongated domains that associate in pairs at acidic pH in the stem region of VWF dimeric bouquets. This correspondence is demonstrated by binding of integrin αIIbβ3 to the fourth module seen in EM, VWC4, which bears the VWF Arg-Gly-Asp motif. The C-terminal cystine knot domain dimerizes end-to-end in a manner predicted by homology to TGF-β and orients approximately perpendicular to the VWC domains in dimeric bouquets. Homologies of domains in VWF to domains in other proteins allow many disulfide bonds to be tentatively assigned, which may have functional implications.
Introduction
von Willebrand factor (VWF) has a central role in hemostasis and thrombosis in the arterial side of the vascular system.1-4 VWF monomers are linked tail-to-tail and head-to-head in VWF concatamers. VWF is a mosaic protein composed of many types of domains (Figure 1). Many of these domains have specific functions in hemostasis; others function in concatamer formation during biosynthesis or give VWF the length and flexibility that enable the bird's nest to elongated conformational transition that activates hemostasis.5
The work described herein builds on early, significant work on the determination of the protein sequence, disulfide connectivity, cDNA sequence, and genomic sequence of VWF.6-14 VWF is one of the largest and most complex mosaic proteins to be characterized. Its domains are the founding members of the von Willebrand A (VWA), von Willebrand C (VWC), and von Willebrand D (VWD) protein families. The original domain designations from the early cDNA cloning papers of VWF (Figure 1A) still predominate in the VWF literature, despite subsequent advances,15 including sequence annotation in protein databases (Figure 1B). In the present study, we update our view of the architecture of the domains within VWF and relate domain structures that are visible by electron microscopy (EM) with sequence repeats and homologies that are detectable by sequence analysis.
Our understanding of the organization and boundaries of domains in VWF remains imperfect. Only the 3 A domains are well characterized and their crystal structures determined.5 Previous chemical assignments of disulfides within VWF were limited to a minority of cysteines in mature VWF that were spaced far enough apart in sequence13,14 (Figure 1A shows assigned disulfides linked by horizontal lines).
EM studies have advanced our understanding of how VWF domains are organized in the acidic conditions of the trans-Golgi and in Weibel-Palade storage granules.5 At the acidic pH of the trans-Golgi, the D1 and D2 domains in the prodomain and D′D3 domain in mature VWF assemble into the helical tubules that characterize Weibel-Palade bodies.16 Earlier in the endoplasmic reticulum, VWF monomers become disulfide linked through their C-terminal cystine knot (CTCK) domains. At the acidic pH of the trans-Golgi, association between the 2 monomers is increased by noncovalent interactions that extend N-terminally from the CTCK domains to the A2 domains, so that the C-terminal two-thirds of the VWF dimer zips up into a dimeric structure, resembling a bouquet of flowers (Figure 1D).17 In dimeric “bouquets,” the closely associated A2, A3, and D4 domains resemble flowers, whereas small domains corresponding to the B and C repeats (Figure 1A) resemble a stem (Figure 1D). However, the small “stem” domains were not enumerated or equivalenced with VWF sequence. The dimeric bouquet structure is pH dependent, so after secretion at the plasma pH of 7.4, it unzips and the dimeric unit is visualized as randomly oriented globules (A2 to D4) connected by thin, flexible strings (the stem region) to a small globule (the CTCK dimer).17 When VWF isolated from plasma is brought to pH 6.2, dimeric bouquets reform as “pendants” on VWF “necklaces.”17
Bork updated VWF annotation by identifying one more VWC repeat, which replaced a portion of D4 and B1 (Figure 1B).15 However, VWC repeats are usually observed in tandem in the protein sequence database, and the assignment of 3 VWC repeats in VWF left 3 intervening gaps, with the Arg-Gly-Asp (RGD) sequence in the middle gap (Figure 1B). Structures have been determined for 2 isolated VWC domains, from collagen IIA18 and from the chordin family member crossveinless 2 in complex with bone morphogenetic protein 2.19
The VWF D domains are now annotated as containing VWD domains, cysteine 8 (C8) domains, and trypsin-inhibitor–like (TIL) domains in the protein family (Pfam) database20 or VWD and TIL domains in the universal protein resource (UniProt) database (Figure 1B). Therefore, the VWD repeat corresponds to a shorter segment than the previously defined D domains (Figure 1A-B). VWD, C8, and TIL repeats show homology to repeats in many proteins beyond VWF. TIL domains have been characterized structurally and have 5 highly conserved disulfide bonds.21,22 Despite these advances, substantial gaps exist between or adjacent to the VWD, C8, and TIL domains in VWF, and these gaps contain many cysteines (Figure 1B), suggesting that domain boundaries may extend further than defined by sequence repeats or that additional domains remain to be identified.
In the present study, we integrated annotation on VWF in databases and knowledge on disulfide linkage and structure of homologous domains with further sequence analysis and tests of domain boundaries by truncation. In combination with EM, this study provides a comprehensive view of the architecture of individual domains and how they are organized within VWF (Figure 1C).
Methods
Sequence alignment
Sequences were aligned with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or PRRN (http://www.genome.jp/tools/prrn/) using progressive pairwise alignment plus iterative refinement. Alignments were in some cases manually adjusted taking into account conserved cysteines and hydrophobic residues.
Constructs and cell culture
VWF fragments with preproprotein numbering were PCR amplified from wild-type human VWF template.17 Constructs D1-D2 (23-763), A3-CK(1683-2813), and D4-CK(1873-2813) in the ET8 vector were as described previously.17 For D1, D4, and D4-VWC fragments, constructs were also cloned into the ET8 vector, with an N-terminal signal peptide and a C-terminal (His)6 tag. All constructs were confirmed by DNA sequencing. VWF constructs were transfected transiently into HEK 293S N-acetylglucosaminyltransferase I-deficient (GntI−) cells23 using polyethylenimine.24 The headpiece fragment containing the β-propeller domain in αIIb and PSI to I-EGF1 domains in β3 was expressed and prepared exactly as described previously.25
The expression levels of constructs in supernatants were assayed by Western blotting using rabbit anti-His polyclonal Abs (Delta Biolabs) followed by HRP-conjugated anti–rabbit whole Ab and enhanced chemiluminescence (GE Healthcare). Images were acquired with an LAS-4000 imager (Fujifilm).
Protein purification
Purification of VWF fragments was as described previously.17 Briefly, proteins were purified using Ni-NTA and Mono-Q columns. Superdex 200 10/300 GL or Sepharose 6 10/300 GL gel filtration (GE Healthcare) was the last step either in 20mM Bis-Tris, pH 6.2, 0.15M NaCl, and 10mM CaCl2 (D1, D4, A3-CK, and D4-CK) or 20mM HEPES, pH 7.4, and 0.15M NaCl (D1-D2). To test complex formation between VWF and integrin, concentrated stock proteins (10 mg/mL for VWF D4-CK and 1 mg/mL for the αIIbβ3 headpiece) at a 1:2 VWF dimer: integrin molar ratio were diluted 5-fold in either 20mM Bis-Tris, pH 6.2, or 20mM HEPES, pH 7.4, buffer containing 0.1M NaCl, 5mM MnCl2, and 0.5mM CaCl2, and loaded onto Superdex 200 10/300 GL (GE Healthcare) in the same buffer. The complex peak fraction at pH 6.2 was immediately subjected to negative-stain EM.
EM and image processing
EM grid preparation and data collection were as described previously.17,26 Briefly, samples fresh from gel filtration were stained on grids with 0.75% (wt/vol) uranyl formate,27 and images were collected on a Tecnai T12 electron microscope (FEI). Particles were picked with BOXER in EMAN,28 and image processing was carried out with SPIDER.29 Details of particle numbers class averaging and the radius for averaging are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and all class averages are shown in supplemental Figure 1. We used a special script to bring subregions of particles into better focus by eliminating the contribution of other regions to the image alignment and class averaging procedure. Particles were first aligned using a large mask including the entire particle, resulting in a stack of aligned particles. A vector was then specified between the old center of alignment and a new center of alignment, and a smaller radius was specified for alignment and averaging.
Results
Subdivision of the VWF D domains
VWF D domains were subdivided into VWD, C8, and TIL modules in Pfam (Figure 1B). We improved boundary assignment (Figure 1C) by applying the principles that in extracellular proteins, most disulfide bonds occur within domains, limited protease digestion under nondenaturing conditions selectively occurs at flexible junctions between domains, and that domains should be contiguous and not leave unexplained cysteine-containing sequences between them.
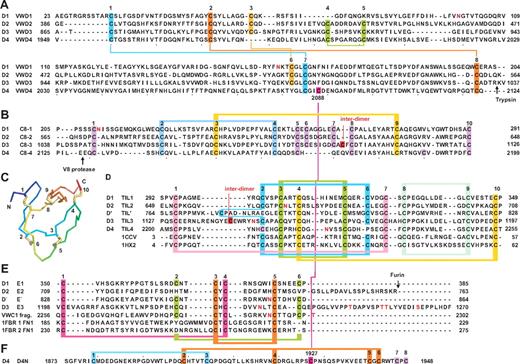
VWF domain annotations. Cysteines are shown as vertical lines and are connected for chemically defined disulfide bonds.13,14 N- and O-linked glycans are shown as closed and open lollipops, respectively.12 (A) Commonly used domain assignment in the literature.2 (B) Annotation in protein databases according to regions of sequence homology. (C) Domain architecture assigned in the present study. Additional disulfide bonds are from predictions described in the text. Boundaries of D assemblies are shown as vertical dashed lines. (D) Dimeric bouquets in acidic pH as found in the Golgi. Organization of the A1-CTCK region is from Zhou et al.17 Organization of the D1-A1 region is predicted based on data of Huang et al.16
VWF domain annotations. Cysteines are shown as vertical lines and are connected for chemically defined disulfide bonds.13,14 N- and O-linked glycans are shown as closed and open lollipops, respectively.12 (A) Commonly used domain assignment in the literature.2 (B) Annotation in protein databases according to regions of sequence homology. (C) Domain architecture assigned in the present study. Additional disulfide bonds are from predictions described in the text. Boundaries of D assemblies are shown as vertical dashed lines. (D) Dimeric bouquets in acidic pH as found in the Golgi. Organization of the A1-CTCK region is from Zhou et al.17 Organization of the D1-A1 region is predicted based on data of Huang et al.16
We subdivided the D1, D2, and D3 domains into VWD, C8, TIL, and E modules (Figure 2). D′ contains the TIL and E subdomains (Figure 1C). The E module newly identified here is an extra segment that follows TIL in D1, D2, D3, and D′. We named modules after the D domain in which they were present: for example, the VWD1, C8-1, TIL1, and E1 modules in D1.
Sequence and structural modules present in D assemblies. (A) VWD modules. (B) C8 modules. (C) Structure of a TIL domain (PDB code 1CCV) in a rainbow representation (blue N to red C) with cysteines numbered and disulfide bonds shown in yellow. (D) TIL modules. (E) E modules. (F) The unique D4N module. Cysteines characteristic of each type of module are numbered above the sequences and are color coded and connected with colored lines as follows: in panel A, all disulfides are defined chemically in 1 or both of D3 and D4; panels B and D through F, disulfide bonds defined chemically in D′D3 or D4 are shown as horizontal connecting lines above the alignments, except in 4 cases of disagreement with homology-defined disulfides (see supplemental Methods); and disulfides suggested by homology are shown as horizontal connecting lines below the alignments. Cleavage sites referred to in the text are marked with arrows. N- and O-linked glycosylation sites12 are shown in red.
Sequence and structural modules present in D assemblies. (A) VWD modules. (B) C8 modules. (C) Structure of a TIL domain (PDB code 1CCV) in a rainbow representation (blue N to red C) with cysteines numbered and disulfide bonds shown in yellow. (D) TIL modules. (E) E modules. (F) The unique D4N module. Cysteines characteristic of each type of module are numbered above the sequences and are color coded and connected with colored lines as follows: in panel A, all disulfides are defined chemically in 1 or both of D3 and D4; panels B and D through F, disulfide bonds defined chemically in D′D3 or D4 are shown as horizontal connecting lines above the alignments, except in 4 cases of disagreement with homology-defined disulfides (see supplemental Methods); and disulfides suggested by homology are shown as horizontal connecting lines below the alignments. Cleavage sites referred to in the text are marked with arrows. N- and O-linked glycosylation sites12 are shown in red.
D4, which is followed by the VWC1 domain, lacks an E segment (Figure 1C). D4 also contains a unique segment between the A3 and VWD4 domains that we designated D4N (Figures 1C, 2F). D4N is linked by a disulfide to VWD4 (Figure 2F). Otherwise, it has 8 cysteines, 2 of which are chemically assigned in a 1-3 linkage.13
VWD domains
Two pairs of disulfide bonds chemically assigned in VWD3 and VWD4, 1-7 and 2-8, are conserved in VWD1 and VWD2 (Figure 2A).13 (Here, cysteines are numbered above sequence alignments and 1-7 refers to the disulfide bond between the corresponding cysteines.) Assignment of the 4-5 disulfide in VWD3 and VWD413 was confirmed by its specific absence in VWD1. Similarly, the 3-6 disulfide assigned in VWD3 was confirmed by its specific absence in VWD4 (Figure 2A). The boundaries between VWD3 and C8-3 were confirmed by limited trypsin digestion (site marked in Figure 2A).13 Fragments containing VWD4 and C8-4 were similarly separable after limited V8 protease digestion17,30 (site marked in Figure 2B). All VWD subdomains contain an even number of cysteines, except VWD4, C2088 of which is disulfide-bonded to C1927 of D4N13 (Figure 2A,F).
C8 domains
The C8 domain (Pfam PF08742) is found in proteins including crossveinless 2, zonadhesin, and mucins, often with N-terminal VWD and C-terminal TIL domains. The 2-4 and 3-9 disulfides chemically demonstrated in C8-313 involve conserved C8 cysteines. C8-1 and C8-2 contain 10 cysteines, all of which may be present in intradomain disulfides, as may be 10 of the 11 cysteines in C8-3 and C8-4. Cys-1099 in C8-3 participates in a head-to-head interdimer disulfide bond in VWF.31 The odd number of cysteines in both C8-4 and TIL4 (Figure 2B-C) may indicate one disulfide between them.
TIL domains
The TIL domain (PF01826) may be associated with VWD and C8 subdomains, or by itself in protease inhibitors, including potent anticoagulants in Caenorhabditis elegans.32 Structures show conserved disulfide linkages (Figure 2C-D), which agree with chemical assignment of the 2-6, 3-5, and 8-9 disulfides in TIL′.13,21,22,32 Homology further identified 1-7 and 4-10 disulfides in VWF TIL domains (Figure 2D). In addition, Cys-1142 in TIL-3 participates in an interdimer VWF disulfide.31
E repeats
E repeats follow each TIL domain except TIL4 (Figure 1C). Each E repeat has either 4 or 6 cysteines. VWC domains structurally contain 2 tandem domains, the first of which is similar to the FN1 module of fibronectin.18,19 The cysteines and certain other residues in E repeats aligned well to the first tandem subdomain in VWC and FN1 domains (Figure 2E); the VWC domain following TIL4 might make up for the lack of a following E repeat. The 1-4 and 3-5 disulfide linkages drawn in Figure 2E are based on those in structurally known VWC and FN1 domains.18,19,33 The remaining 2 cysteines in E repeats would thus be predicted to be linked to 2-6 in E repeats, in agreement with the close proximity of the corresponding residues in FN1 domains33 and the specific absence of these cysteines in E2 (Figure 2E). Supplemental Methods detail a minority of 4 disulfides in C8, TIL, and E domains that are discrepant when assigned chemically and by homology.
D domains contain interdependent lobes
Expression of fragments in HEK 293 cells, followed by Western blotting, was used to identify the minimal units required in D1 and D4 for secretion (Figure 3A-B). A D1 fragment (23-385) containing VWD1, C8-1, TIL1, and E1 was successfully expressed in the absence of D2 (Figure 3B lane 2). Deletion of the E1 segment in the 23-349 fragment abolished expression (Figure 3B lane 1). Expression using previously defined boundaries of the D4 domain (1949-2299) was unsuccessful (Figure 3B lane 9).
Expression and visualization of D assemblies. (A) D1 or D4 constructs. Cysteines are shown in vertical lines, and are connected for known and predicted disulfide bonds. (B) Expression of constructs by transient HEK293S GntI− transfectants as measured by Western blotting with anti-His tag of equal quantities of supernatants subjected to SDS 10% PAGE. (C) Superdex 200 gel-filtration of purified D1 (23-385) and D4(1873-2255) in 20mM Bis-Tris, pH 6.2, 0.15M NaCl, and 10mM CaCl2. Elution positions of calibration proteins are marked. (D) SDS-PAGE of D1 and D4 constructs with and without endoglycosidase H treatment stained with Coomassie blue. (E) Raw EM image of D4(1873-2255) in 20mM Bis Tris, pH 6.2, 0.15M NaCl, and 10mM CaCl2. (F) Representative class averages of D4(1873-2255). (G) Comparison between class averages of D4-CK head (aligned with a radius of 12 nm) and D4 (1873-2255). (H) Representative class averages of D1 (23-385) monomers at pH 6.2. (I) Raw EM image of D1-D2 (23-763) in 20mM HEPES, pH 7.4, and 0.15M NaCl. (J) Representative class averages of D1-D2. Scale bars represent 50 nm in raw images and 5 nm in class averages.
Expression and visualization of D assemblies. (A) D1 or D4 constructs. Cysteines are shown in vertical lines, and are connected for known and predicted disulfide bonds. (B) Expression of constructs by transient HEK293S GntI− transfectants as measured by Western blotting with anti-His tag of equal quantities of supernatants subjected to SDS 10% PAGE. (C) Superdex 200 gel-filtration of purified D1 (23-385) and D4(1873-2255) in 20mM Bis-Tris, pH 6.2, 0.15M NaCl, and 10mM CaCl2. Elution positions of calibration proteins are marked. (D) SDS-PAGE of D1 and D4 constructs with and without endoglycosidase H treatment stained with Coomassie blue. (E) Raw EM image of D4(1873-2255) in 20mM Bis Tris, pH 6.2, 0.15M NaCl, and 10mM CaCl2. (F) Representative class averages of D4(1873-2255). (G) Comparison between class averages of D4-CK head (aligned with a radius of 12 nm) and D4 (1873-2255). (H) Representative class averages of D1 (23-385) monomers at pH 6.2. (I) Raw EM image of D1-D2 (23-763) in 20mM HEPES, pH 7.4, and 0.15M NaCl. (J) Representative class averages of D1-D2. Scale bars represent 50 nm in raw images and 5 nm in class averages.
However, altering both the N and C-terminal boundaries enabled expression of the 1873-2255 fragment containing D4N, VWD4, C8-4, and TIL4 (Figure 3B lane 4). Omission of D4N in 1947-2255 and 1949-2255 fragments abolished expression.
The well-expressed VWD1-E1 and D4N-TIL4 fragments, which we termed the D1 and D4 assemblies (Figure 1C), were purified and examined by gel-filtration (Figure 3C) and SDS-PAGE, followed by Coomassie blue staining (Figure 3D). Both D1 and D4 assemblies gave symmetric single peaks in gel-filtration analysis (Figure 3C). They eluted earlier than expected for globular domains of calculated polypeptide masses of 45.8 kDa for D1 and 44.9 kDa for D4 (Figure 3), which is consistent with the presence of multiple lobes described in the following paragraph. Treatment with endoglycosidase H of material expressed in GntI− cells reduced the Mr in SDS-PAGE from 48.1 to 44.1 kDa and from 49.9 to 49.0 kDa for D1 and D4 assemblies, respectively, consistent with presence of 3 and one consensus NX(T/S) glycosylation sequons, respectively (Figure 3D). The size in reducing and nonreducing SDS-PAGE, and lack of disulfide-linked multimers (Figure 3B,D) was consistent with both assemblies being monomeric. The greater shift in migration between reducing and nonreducing SDS-PAGE for D4 than D1, seen in both Figure 3B and D, is consistent with the chemically determined, long-range disulfide bond between the D4N and VWD4 domains.
Negative-stain EM followed by picking of 3600-6000 particles and particle alignment and class averaging (supplemental Table 1 and supplemental Figure 1) showed that the D4 assembly consists of 3 or 4 lobes that associate closely, justifying the term “assembly” (Figure 3E,F). Some class averages of D4-CK dimeric bouquets also showed multiple lobes in D4 (Figure 3G top) and gave high cross-correlation scores with isolated D4 assembly (Figure 3G bottom). Class averages of the VWD1-E1 fragment showed that it also was an assembly of 3-5 closely associated lobes (Figure 3H). Furthermore, the D1-D2 prodomain consists of 2 separate assemblies joined by a thin connection, with each assembly containing multiple lobes (Figure 3I-J).
D assembly overview
In conclusion, EM provides a bird's-eye view of the D regions as assemblies, composed of smaller modules or lobes that associate in specific ways. This agrees with the definition of multiple sequence modules termed VWD, C8, TIL, E, and D4N within D assemblies. Visualization of either 3 or 4 lobes in EM is likely a consequence of different orientations of the assemblies on the substrate. Cooperative association between modules within assemblies is demonstrated by the successful expression of the D1 assembly as defined, but not when its C-terminal E1 module was omitted. Similarly, expression of the D4 assembly was successful when all 4 modules were present, but not when its N-terminal D4N or C-terminal TIL4 modules were omitted.
Because the 4 modules in D assemblies differ in size, with VWD being so much larger than the others, they almost certainly do not correspond 1:1 with the 4 similarly sized densities per D region often seen in EM. We propose that the VWD domain is divided into N- and C-terminal lobes that are bridged by the 1-7, 2-8, and 3-6 long-range disulfides. This proposal is consistent with the richness of Gly and hydrophilic residues adjacent to these cysteines, as opposed to the hydrophobic residues that would be expected if these cysteines were in the core of a domain.
The D4 assembly uniquely contains a D4N module. EM of dimeric bouquets shows a prominent hook region bridging crescent-shaped D4 domains at their A3-proximal end; this bridging region might correspond to D4N.17 The D′D3 assembly is unique in containing the “extra” TIL′ and E′ modules (Figure 1). Class averages of D′D3-A1 dimers show a horn-like protrusion from a D3 assembly that might correspond to TIL′ and E′.17
It is unclear whether any of the VWD, C8, TIL, or E segments in D domains have specific or separable functions. However, mutations in VWF that specifically decrease binding to factor VIII (type 2N von Willebrand disease) map to both the TIL′ and E′ segments, suggesting a direct role in binding factor VIII.34 Whether any of the TIL domains in VWF have protease inhibitor activity similar to other TIL family members21,22,32 would be interesting to determine.
VWC domains
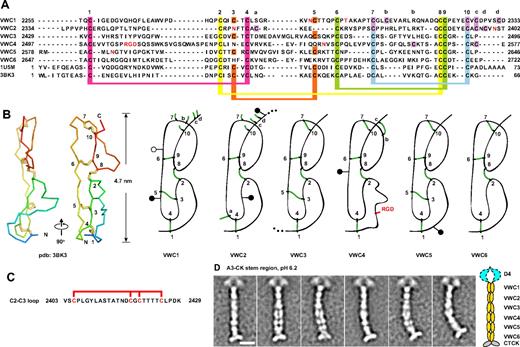
Three domains in VWF, previously termed C1, C2, and C3 (Figure 1B), andC1, C3, and C5 herein (Figure 1C), are VWC domains by sequence homology.15 Three intervening regions are similar in length and sequence to VWC domains, which we designated the VWC-like C2, C4, and C6 domains (Figures 1C, 4A). Structures of VWC domains in other proteins18,19 show 2 subdomains linked in tandem in an extended orientation, define the linkage of 10 conserved cysteines (Figure 4A-B), and enable disulfide assignment in VWF VWC domains by homology (Figure 4A). Confidence in C2, C4, and C6 as VWC domains is increased by the predicted sharing of 4 disulfide bonds with VWC domains, and the specific absence of a pair of cysteines, 3 and 5, that are disulfide linked in VWC (Figure 4A).
VWC repeats. (A-B) VWC sequence and structure. VWC sequences in VWF are aligned with one another and with structurally characterized VWC domains (A) and shown schematically in relation to structure (B). Conserved cysteines (numbered) and extra cysteines (lettered) are located in both panels A and B. N- and O-linked glycosylation sites are in red (A) and shown as closed and open lollipops, respectively (B). (C) Segment between VWC2 and VWC3 containing 2 local disulfide bonds.13 (D) Representative class averages of the A3-CK dimeric bouquet, aligned centering on the fourth globule in the stem region, with a radius of 20 nm. Scale bar represents 10 nm.
VWC repeats. (A-B) VWC sequence and structure. VWC sequences in VWF are aligned with one another and with structurally characterized VWC domains (A) and shown schematically in relation to structure (B). Conserved cysteines (numbered) and extra cysteines (lettered) are located in both panels A and B. N- and O-linked glycosylation sites are in red (A) and shown as closed and open lollipops, respectively (B). (C) Segment between VWC2 and VWC3 containing 2 local disulfide bonds.13 (D) Representative class averages of the A3-CK dimeric bouquet, aligned centering on the fourth globule in the stem region, with a radius of 20 nm. Scale bar represents 10 nm.
There may be interactions between each pair of tandem VWC domains. An uneven number of cysteines can be accommodated in VWC1 and VWC2, because Cys a in VWC2 is predicted to disulfide bond across the VWC1-VWC2 junction to either cysteine b, c, or d in VWC1 (Figure 4B). The C1-C2 pair is separated from C3-C4 by a unique Ser- and Thr-rich segment of 26 residues containing 4 cysteines that are internally disulfide bonded13 (Figure 4C). The last 9 residues of C3 and C5 are identical and the first 3 residues of C4 and C6 are almost identical7 (Figure 4A), suggesting a conserved interaction at the C3-C4 and C5-C6 junctions. Some of the extra cysteines, labeled a, b, c, and d (Figure 4A), may be tentatively assigned based on VWC structure (Figure 4B).
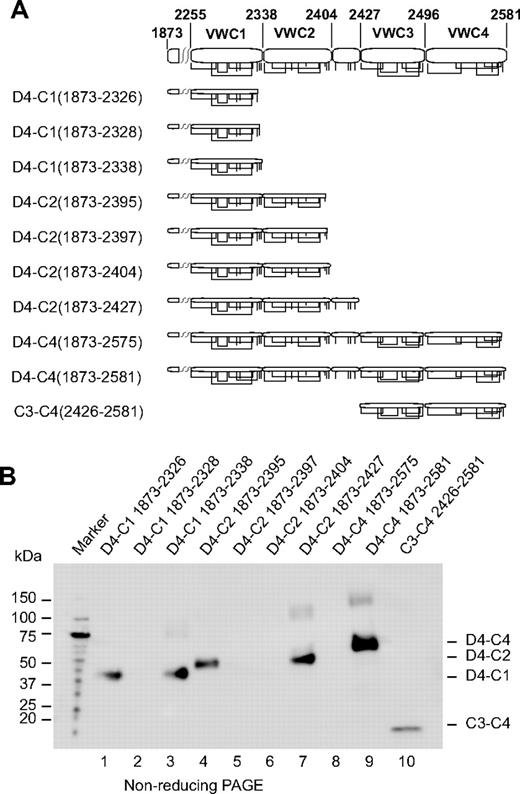
Truncations in the C-repeat region (Figure 5A) were tested for secretion (Figure 5B). A truncation that began in D4 and extended through the predicted end of VWC1 (D4-C1 1873-2338) was expressed well (Figure 5B lane 3), whereas truncations lacking the last 1 or 2 cysteines were expressed less well or undetectably (Figure 5B lanes 1 and 2, respectively). A truncation extending to Cys 10 of VWC2 (D4-C2 1873-2395) was expressed better (Figure 5B lane 4) than truncations containing the last 1 (D4-C2 1873-2397) or 2 unassigned cysteines (D4-C2 1873-2404) (Figure 5B lanes 5 and 6). A fragment including the segment containing 4 Cys following VWC2 (D4-C2 1873-2427) was also well expressed (Figure 5B lane 7). A segment extending to the predicted end of VWC4 (D4-C4 1873-2581) was very well expressed (Figure 5B lane 9); however, a construct missing the last Cys of VWC4 was not expressed at all (Figure 5B lane 8). Furthermore, the predicted boundaries N-terminal to VWC3 and C-terminal to VWC4 were confirmed by successful expression of the C3-C4 (2426-2581) fragment (Figure 5B lane 10).
Expression of VWC constructs. (A) Diagram of constructs. Cysteines are shown as vertical lines and are connected according to predicted linkages. (B) Anti-His tag Western blot. Supernatants (equal volumes) from HEK293S GnTI− transient transfectants were subjected to nonreducing SDS 4%-15% gradient PAGE and Western blotting.
Expression of VWC constructs. (A) Diagram of constructs. Cysteines are shown as vertical lines and are connected according to predicted linkages. (B) Anti-His tag Western blot. Supernatants (equal volumes) from HEK293S GnTI− transient transfectants were subjected to nonreducing SDS 4%-15% gradient PAGE and Western blotting.
Ganderton et al35 have independently recognized 3 VWC-like domains that were termed joiner domains. However, we have recognized residues 2403-2429 as a separate element (Figure 4C), and therefore the alignment of their C1C2 joiner domain, which includes these residues, differs from the alignment of our VWC2 domain. They successfully expressed fragments corresponding in our nomenclature to C1-CK, C1-C5, and C3 and found that one cysteine residue in the C1-CK fragment, C2453, was only 25% disulfide bonded.35
VWC stem modules and the binding site for integrin αIIbβ3
EM on the dimeric bouquet of VWF has shown that the region with VWC domains is stem-like.17 To obtain greater detail in this region, the class-averaging procedure was altered to use a smaller radius centered on the middle of the stem for alignment and averaging (see “Methods”). The resulting class averages of A3-CK dimers at pH 6.2 sharpen resolution in the stem while allowing A3 (not shown) and D4 to blur (Figure 4D). The averages are consistent with 6 sets of paired domains in the stems lying either beside or on top of one another in the stem.
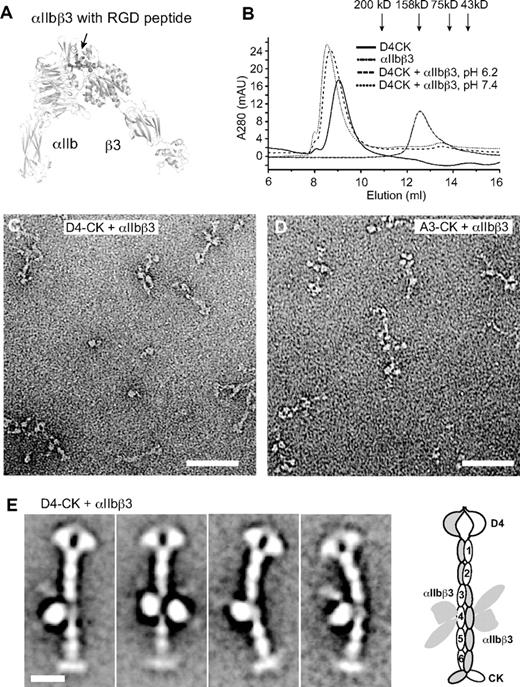
The integrin αIIbβ3–binding site in VWF was used as a landmark to confirm the register between modules defined by sequence and seen in EM. The RGD sequence recognized by integrin αIIbβ336 was in VWC4 (Figure 4A). The headpiece of integrin αIIbβ3 (Figure 6A) complexed in Mn2+ with D4-CK dimer as shown by earlier elution in gel filtration than either component (Figure 6B). Complexes were obtained at both pH 6.2, where dimers are zipped up, and at pH 7.4, where they are unzipped (Figure 6B). EM at pH 6.2 revealed integrin heterodimers bound to VWF dimers, along with some dissociation that occurred either during gel filtration or adsorption to EM grids (Figure 6C-D). Class averages show either 1 or 2 integrins bound per dimer (Figure 6E). Binding of αIIbβ3 to the fourth stem module demonstrates its equivalence to the RGD-bearing VWC4 module (Figure 6E).
Binding of the integrin αIIbβ3 headpiece to the VWF D4-CK fragment. (A) Overall shape of the open αIIbβ3 headpiece (PDB code 2VDR and 3FCS), including the β-propeller and thigh domains in αIIb and PSI, hybrid, βI, and I-EGF1 domains in β3 subunit. Arrow shows the position of the RGD-binding site. (B) Superdex 200 gel filtration of VWF D4-CK, αIIbβ3 headpiece, and their complex in 5mM MnCl2, 0.5mM CaCl2, and 0.1M NaCl and in 20mM buffer of either Bis-Tris, pH 6.2, or HEPES, pH 7.4. (C) EM image of the complex of D4-CK with the αIIbβ3 headpiece at pH 6.2. (D) EM image of the complex of A3-CK with the αIIbβ3 headpiece at pH 6.2. (E) Representative class averages of complex of D4-CK with the αIIbβ3 headpiece. A schematic of integrin binding to VWF is shown to the right. Scale bars represent 50 nm in images and 10 nm in class averages.
Binding of the integrin αIIbβ3 headpiece to the VWF D4-CK fragment. (A) Overall shape of the open αIIbβ3 headpiece (PDB code 2VDR and 3FCS), including the β-propeller and thigh domains in αIIb and PSI, hybrid, βI, and I-EGF1 domains in β3 subunit. Arrow shows the position of the RGD-binding site. (B) Superdex 200 gel filtration of VWF D4-CK, αIIbβ3 headpiece, and their complex in 5mM MnCl2, 0.5mM CaCl2, and 0.1M NaCl and in 20mM buffer of either Bis-Tris, pH 6.2, or HEPES, pH 7.4. (C) EM image of the complex of D4-CK with the αIIbβ3 headpiece at pH 6.2. (D) EM image of the complex of A3-CK with the αIIbβ3 headpiece at pH 6.2. (E) Representative class averages of complex of D4-CK with the αIIbβ3 headpiece. A schematic of integrin binding to VWF is shown to the right. Scale bars represent 50 nm in images and 10 nm in class averages.
VWC overview
In the dimeric bouquet, the 6 tandem VWC domains have a highly extended conformation. Each VWC domain pairs with its counterpart in the other monomer. Pairing between identical VWC modules was confirmed by binding of 2 αIIbβ3 headpiece fragments to paired VWC4 modules.
Crystal and nuclear magnetic resonance spectroscopy structures of single VWC modules showed an N- to C-length of 4.7-5.0 nm, and flexibility between the 2 tandem, extended, disulfide-linked subdomains in each VWC module.18,19 Multiplied by 6, this gives 28-30 nm, close to the measured length of the stem region in EM of approximately 32 nm. These comparisons suggest that in dimeric bouquets, the long axes of the VWC domains are aligned with the stem, consistent with visualization of elongated, paired globules in some EM class averages (Figure 4D).
Close packing between VWC domains in the stem suggests highly specific pairwise interactions. VWC1-VWC1 and VWC2-VWC2 packings appeared to be particularly tight, because these pairs were always seen as single globules in class averages, whereas the other pairs were seen as either a single merged or 2 side-by-side globules, depending on the class average. The distance along the stem was also lesser for the VWC1-VWC1 and VWC2-VWC2 pairs than the others, suggesting a greater crossing angle for their N to C axes. This transition in length per VWC module occurs at the insertion position of the unique 4-cysteine–containing 2403-2429 segment between VWC2 and VWC3, which is too small to be accounted for by separable density in our EM class averages. The VWC domains in dimeric bouquets appear to exhibit some type of twist of each monomer around the dyad axis.
Whereas most easily visualized in EM in dimeric bouquets at the acidic pH characteristic of Weibel-Palade bodies, the 6 tandem VWC modules are most functionally important at neutral pH in plasma, where they contribute great length and flexibility to VWF.17 Compared with immunoglobulin superfamily domains of 90-100 residues and 4 nm in length, the VWC domains of VWF have fewer residues (on average, 74), but extend further, approximately 5 nm per module. Moreover, the flexibility seen in nuclear magnetic resonance spectroscopy18 and lack of a hydrophobic core are expected to give the VWC domains in VWF a rope-like character, with bending occurring throughout the length of VWC domains as well as at their junctions. In contrast, tandem repeats of larger domains such as IgSF domains show bending only at interdomain junctions.
VWC domains are highly represented in matrix proteins that regulate growth factor responses by binding to bone morphogenetic proteins in the TGF-β family.15,19 von Willebrand disease can be associated with vascular malformations and altered growth characteristics of patient endothelial cells.37 Therefore, it will be interesting to determine whether the VWC domains of VWF have a role in the regulation of angiogenesis by bone morphogenetic proteins.38
The CTCK domain
The CTCK dimer forms a tee-shaped base for the dimeric bouquet (Figure 4D). The long axes of the 2 VWF CTCK domain monomers are approximately perpendicular to the long axis of the 6 VWC domain tandems. The 2 CTCK domain monomers link at one end at an angle of approximately 130° (Figure 4D).
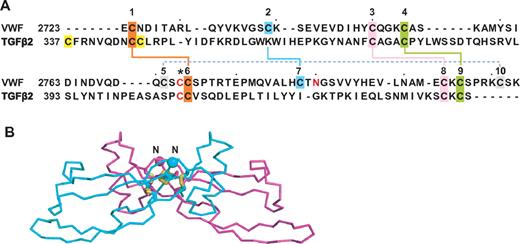
The cystine knot is a motif found in diverse proteins in which 1 disulfide passes between 2 disulfides formed by cysteines a few residues apart in each of 2 β-strands that they bridge.8 Cystine-knot cytokines, including PDGF, nerve growth factor, and TGF-β, are structurally related to one another, but differ in position of the cysteine that dimerizes 2 monomers.8 CTCK domains, always found at the C-terminus of extracellular proteins, include Norrie disease protein, mucins, connective tissue growth factor, and VWF.8 Cystine knot cytokines and CTCK domains are sufficiently similar to enable prediction of the disulfide linkage in CTCK domains based on alignment with TGF-β14 (Figure 7A). Chemical assignment confirmed the predicted linkage of 8 of the 11 cysteines in the VWF CTCK, leaving undefined the linkage of 3 others, including the one(s) mediating dimerization.14 Among cystine knot cytokines, TGF-β monomers are disulfide-linked head-to-head with their long axes anti-parallel. In contrast, PDGF monomers are linked both head-to-head and tail-to-tail, so their long axes are parallel. Our EM results support the head-to-head, anti-parallel TGF-β–like dimerization model,8,14 and thus dimerization through a cysteine that aligns with the dimerizing cysteine in TGF-β8 (Figure 7A asterisk). The overall shape of the TGF-β dimer seen in crystal structures (Figure 7B) is consistent with the shape and orientation between the 2 CTCK monomers seen in EM of VWF dimeric bouquets (Figure 4D). The tee-like orientation between VWC and CTCK domains in VWF may be a common feature of the many proteins that have VWC tandems terminated by CTCK domains.8
CTCK domain. (A) Sequence alignment of VWF CTCK domain and TGF-β2 taken from Meitinger et al.8 Disulfides found both chemically14 and predicted8 or only predicted8 are shown with solid or dashed lines, respectively, and the cysteine predicted to form the interchain disulfide is shown with an asterisk. (B) Ribbon diagram of TGF-β2 crystal structure.40 The most N-terminal region that does not align to VWF CTCK is deleted. The new N-terminus is shown as a Cα sphere.
CTCK domain. (A) Sequence alignment of VWF CTCK domain and TGF-β2 taken from Meitinger et al.8 Disulfides found both chemically14 and predicted8 or only predicted8 are shown with solid or dashed lines, respectively, and the cysteine predicted to form the interchain disulfide is shown with an asterisk. (B) Ribbon diagram of TGF-β2 crystal structure.40 The most N-terminal region that does not align to VWF CTCK is deleted. The new N-terminus is shown as a Cα sphere.
Discussion
The entire sequence of VWF is accounted for herein, except for the well-characterized VWA domains. VWA domains are large and globular, with hydrophobic cores, whereas the remaining modules are small and predicted to lack hydrophobic cores, accounting for their richness in disulfide bonds. Previous chemical definitions of disulfide bonds are shown in Figure 2A (VWD), and by connections over the alignments shown in Figure 2B, D, and F (the C8, TIL, and D4N modules, respectively).13 Prediction by homology and specific absences of disulfides enables identification of additional disulfides in the TIL domains, and identification of disulfides for the first time in the E and VWC domains, as shown by connections below the alignments in Figure 2D and E and Figure 4A. Previous predictions in the CTCK domain8 have been confirmed14 (Figure 7A). The disulfides newly predicted here are useful for guiding further experimentation, but should be considered tentative until confirmed chemically or structurally.
In the present study, we have predicted all modules and their boundaries within VWF and have shown for the first time that the D domains are assemblies composed of smaller modules. The largest of these modules is the VWD module, which while having a sufficient number of residues to contain a hydrophobic core, may nonetheless contain smaller units that are linked by disulfide bonds. The boundaries of the D assemblies have been confirmed by expression and EM experiments. The boundaries between VWC modules C1 and C2, C2 and C3, and C4 and C5, as well as between D4 and C1, were also confirmed by expression; slight ambiguity remains about the C1-C2 boundary. However, we suggest that there may be significant interactions between the C1-C2, C3-C4, and C5-C6 module pairs, and their boundaries may not be well defined, even at the structural level. Our prediction of 6 VWC modules was confirmed by EM, as was the register between the tandem modules seen in EM and sequencing studies. Proteins were expressed here in GntI− cells, which results in high mannose N-linked carbohydrates and no effect on O-linked carbohydrates.23 This facilitates checking glycoproteins for N-glycans using endoglycosidase H and for crystallization and has no deleterious effects on structure. Each of the D1, D2, and D′D3 domains are required for the formation of Weibel-Palade bodies and intracellular storage.16,39 Secretion from mammalian cells requires passage through endoplasmic reticulum quality-control checks and is an excellent surrogate for correct folding. All expressed (ie, secreted) constructs reported herein were cysteine rich yet showed little dimer or multimer formation in nonreducing SDS-PAGE or in gel-filtration, again supporting correct folding. Furthermore, the D1 construct reacted with 2 mAbs to VWF (not shown) and the individually expressed D1 and D4 assemblies were shown by EM to be very similar in appearance to these assemblies in the D1-D2 propeptide and in dimeric VWF constructs, respectively.
Overall, the results of the present study provide important information about the structure of the domains within VWF, both when assembled in helices and zipped up in dimeric bouquets during storage in Weibel-Palade bodies and after secretion into plasma, when the prodomains dissociate and the dimeric bouquets unzip. In plasma, VWF concatamers are highly flexible and flow-influenced transitions between compact and elongated conformations appear to regulate hemostasis.5 Length is dominant over mass in determining the hydrodynamic forces acting on VWF, which in turn regulate VWF function.5 Among modules in VWF, the VWC domains contribute the most to this length and also confer flexibility that is undoubtedly important in the rapid transition between the compact, birds' nest, and elongated conformations of VWF in flow.5
The presence of 3-6 VWD, C8, TIL, E (or FN1-like), VWA, and VWC modules per VWF monomer again emphasizes the mosaic nature of VWF. Only the D4N module, the 4-cysteine module between the C2 and C3 modules, and the CTCK domain occured once per monomer. The only known specific functional roles of the cysteine-rich VWF modules, aside from regulating dimerization and concatamerization, are binding factor VIII and integrin αIIbβ3 (Figure 1). However, homologies described herein of the TIL and VWC domains suggest that it could be interesting to determine their roles as protease inhibitors and growth factor regulators, respectively.
The VWD, C8, TIL, E, and D4N units in VWF are organized in specific ways into compact assemblies that require both the most N- and C-terminal modules for optimal secretion and, presumably, folding. Only in a few cases are modules that neighbor one another in sequence linked by long-range disulfide bonds. Elongational force exerted across D assemblies will pry the N-terminal and C-terminal modules away from one another, open up the assembly, and possibly expose cryptic binding sites.
The overview of VWF architecture described herein allowed us to associate each of 6 VWC repeats and 1 CTCK domain present in the VWF sequence with specific densities of dimeric bouquets visible using EM. Multiple modules present in the sequence of each D assembly were also associated with the presence of multiple lobes seen in EM. In addition to these insights, our annotation and structural studies provide a roadmap for further work on the function and structure of VWF.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Zongli Li and Mr Yang Chen for the script for recentering particle averaging. T.W. is an investigator in the Howard Hughes Medical Institute.
This work was supported by the National Institutes of Health (grant HL-103526).
National Institutes of Health
Authorship
Contribution: Y.-F.Z. prepared the constructs, designed and performed the experiments, and wrote the manuscript; E.T.E. performed the EM experiments; J.Z. and C.L. prepared the constructs, designed and performed the experiments, and discussed the manuscript; T.W. discussed and supervised the EM experiments and strategy and wrote the manuscript; and T.A.S. designed the overall experimental approach, supervised the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Springer, 3 Blackfan Circle, Boston, MA 02115; e-mail: springer@idi.harvard.edu.