Abstract
Abstract 1329
De novo CpG methylation is catalyzed by two enzymes (DNMT3A and DNMT3B), while DNMT1 is responsible for maintenance methylation during cell replication. DNMT3L, a catalytically inactive protein, interacts with and influences DNMT3A and DNMT3B target preference and methylation kinetics. Recurrent mutations in DNMT3A have been found in over 20% of patients with acute myeloid leukemia (AML) and have been associated with poor clinical outcomes (Ley, TJ et al. NEJM, 2010). Greater than 50% of DNMT3A mutations are found at position R882 within the catalytic domain. Because R882H mutations in AML are nearly always heterozygous, because the mutant allele is expressed at the same level as the corresponding WT allele (Ley, TJ et al. NEJM, 2010), and because the mutant enzyme has reduced methyltransferase activity (Yamashita, Y et al. Oncogene, 2010; Holz-Schietinger, C et al. JBC, 2012), it has been suggested that the R882H mutation contributes to leukemogenesis by leading to haploinsufficiency for DNMT3A. However, mice haploinsufficient for Dnmt3a exhibit normal hematopoiesis, while HSPCs lacking Dnmt3a exhibit increased self-renewal and decreased differentiation after serial transplantation (Challen, GA et al. Nat Genet, 2011).
To address this conundrum, we have studied the R882H mutation in a setting that mimics the intrinsic de novo methylation capacity of a typical AML cell. Using expression array and RNA-Seq data from 178 AML patients, we discovered that DNMT3L is not expressed in AML cells, and that DNMT3A is expressed on average 2.3-fold higher than DNMT3B. Interestingly, 92% of AML patients predominantly express inactive splice variants of DNMT3B, regardless of FAB or mutational profile (median ratio of inactive to active DNMT3B transcripts is 3.1:1). Given that the inactive splice variant DNMT3B3 is the most highly expressed isoform in most patients in our cohort, we explored the functional interactions between WT DNMT3A, R882H DNMT3A, and DNMT3B3 using recombinant enzymes made in eukaryotic cells.
In vitro methylation of plasmid DNA (pcDNA3.1) with 3H-SAM using purified recombinant full-length human DNMT3A protein confirmed that the R882H mutation severely reduces the catalytic activity of DNMT3A, resulting in an enzyme with ∼10% of the activity of the WT enzyme. These results were verified by independent in vitro methylation experiments analyzed by bisulfite sequencing, which also revealed that the CpG-flanking sequence preferences of WT and R882H DNMT3A are identical and consistent with the expected “TNCGCY” motif previously described (Wienholz, BL et al. PLoS Genet, 2010). Mixing WT and R882H DNMT3A at equimolar ratios resulted in no significant changes in CpG-flanking sequence preference (compared to WT or R882H enzyme alone; Spearman correlation between WT DNMT3A and WT+R882H DNMT3A = 0.99).
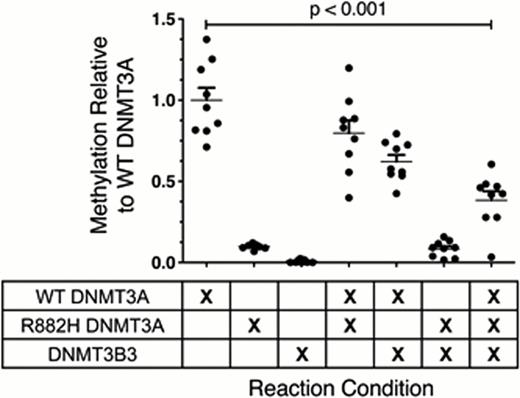
In contrast, mixing WT and R882H DNMT3A at equimolar ratios in a 12-hour methylation assay demonstrated that R882H DNMT3A exerts an inhibitory effect on the catalytic activity of WT DNMT3A in vitro. Instead of increasing net methylation activity by a predicted 10% (summing the activity of the two individual enzymes), R882H DNMT3A led to a 20% reduction in the measured methylation. Similarly, the addition of catalytically inactive DNMT3B3 to WT DNMT3A resulted in a mean decrease in methylation of 38%. Combining equimolar amounts of WT DNMT3A, R882H DNMT3A, and DNMT3B3 led to an additive inhibition of methylation compared to WT DNMT3A alone (62% decrease; p < 0.001; Figure 1). This scenario closely mimics the ratio of these enzymes in AML cells, and our data therefore suggest that the additive inhibitory effects of R882H DNMT3A and DNMT3B3 could severely reduce the total de novo methylation activity of DNMT3A in AML cells. The reduction of enzyme activity below haploinsufficient levels may be important for AML pathogenesis, and these findings provide a mechanism to achieve these levels.
The de novo methyltransferase activity of WT DNMT3A is inhibited by R882H DNMT3A and DNMT3B3. Mixing equimolar amounts of WT DNMT3A, R882H DNMT3A, and DNMT3B3 leads to additive inhibition of methylation by 62% (p < 0.001).
The de novo methyltransferase activity of WT DNMT3A is inhibited by R882H DNMT3A and DNMT3B3. Mixing equimolar amounts of WT DNMT3A, R882H DNMT3A, and DNMT3B3 leads to additive inhibition of methylation by 62% (p < 0.001).
Ley:Washington University: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.