Abstract
Abstract 2874
Although novel treatment options have improved response rates and survival in CLL, the portion of cases with refractory disease remains considerably high and management is challenging. The humanized anti-CD52 monoclonal antibody alemtuzumab is standard treatment in refractory CLL and shows efficacy irrespective of the underlying genomic aberrations or mutation status of TP53. However, only about half of all refractory cases can be explained by TP53 mutation/deletion and treatment remains unsatisfactory.
To elucidate pathogenic and prognostic factors, we performed microRNA expression analyses on a set (n=42) of fludarabine-refractory CLL patients treated within the multicenter phase II CLL2H trial of the GCLLSG (3×30 mg alemtuzumab s.c. per week, max 12 weeks). Profiling was done with the Agilent microRNA microarray platform based on the Sanger miRBase Release 14.0. Statistical analysis was performed with the R software version 2.15.1. Normalization of the arrays was conducted by using the least-variant set (LVS) of genes method and by quantile normalization.
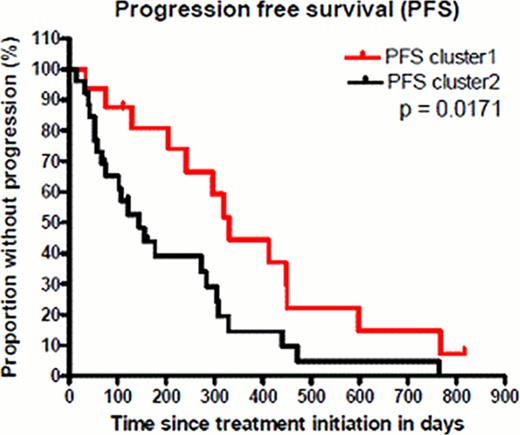
When filtering the expression data based on standard deviation (sd>0.5), we found 131 microRNAs showing greatest variability in expression levels across all arrays. Subsequent unsupervised bootstrap clustering (mean centered; distance: pearson correlation; agglomeration rule: average linkage) resulted in two stable major clusters. Cluster 1 contained more cases with p53 defect which were observed in 56% (TP53 mutation 7/16, 17p deletion 6/16) vs. 27% (TP53 mutation 6/26, 17p deletion 5/26). Cluster 2 was slightly enriched for IGHV unmutated cases in 61% (16/26) vs. 50% (8/16). Median levels of thymidine kinase were 31.6 U/L in cluster 1 and 39.6 U/L in cluster 2, beta2-microglobulin levels were similar in both clusters with a median of 4.3 and 4.4 mg/L. Expression of miR-34a, a microRNA known to execute p53-dependent cell cycle arrest, as well as induction of senescence and apoptosis, was predominantly low in cluster 1. Cases with low miR-34a expression in cluster 1 and without altered p53 (no 17p deletion and/or TP53 mutation) showed a selective upregulation of microRNAs known to be involved in proliferation, neoangiogenesis and inhibition of apoptosis by targeting genes like CDKN1A, E2F, BIM and PTEN. Interestingly, these microRNAs have been reported to be repressed by p53 under physiologic conditions, pointing to alternative mechanisms of p53 pathway inactivation. With regard to treatment response, cluster 1 was significantly enriched for better response (partial response (PR) n=9 (56.25%); stable disease (SD) n=5 (31.25%); progressive disease (PD) n=2 (12.5%)). In contrast cluster 2 (n=26) was dominated by cases with inferior response (PR n=4 (15%); SD n=13 (50%); PD n=9 (35%)). Progression free survival (PFS) was significantly longer in cluster 1 (median 331 days vs. 145 days, p=0.0171, Figure 1). Overall survival was in trend longer in cluster 1 (median 760 days vs. 513 days, p=0.2480).
In conclusion, microRNA expression in fludarabine-refractory CLL shows high variability likely induced by independent mechanisms of resistance. Altered microRNA expression might mimic effects similar to TP53 mutation or 17p deletion even in situations when p53 itself is not functionally disrupted. Of note, unsupervised clustering identified two major clusters that showed a surprising prognostic relevance in fludarabine-refractory CLL treated with alemtuzumab.
Progression free survival of refractory patients treated with alemtuzumab based on major clusters after unsupervised clustering.
Progression free survival of refractory patients treated with alemtuzumab based on major clusters after unsupervised clustering.
Stilgenbauer:Genzyme: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.