Abstract
Abstract  710
710
Genetic alterations play a major role in stratifying prognostic risk groups in chronic lymphocytic leukemia (CLL). Recently, next generation sequencing strategies revealed novel recurrent mutations in genes such as NOTCH1 and SF3B1 in CLL. Correlations have been reported for SF3B1 mutations (SF3B1mut) with deletion 11q and for NOTCH1 mutations (NOTCH1mut) with trisomy 12. Both mutations have been associated with unmutated IGHV, refractory disease and inferior outcome in heterogeneous cohorts outside clinical trials.
We assessed the incidence and prognostic impact of NOTCH1, SF3B1 and TP53 mutations in the CLL2H trial of the GCLLSG, a multicenter phase II trial of subcutaneous alemtuzumab (3 × 30mg/week, max. 12 weeks, n=103, NCT00274976) for fludarabine-refractory CLL patients. Genetic studies were performed in the central reference laboratory of the GCLLSG on 97/103 (94%) trial patients based on availability of DNA. NOTCH1 was analyzed by Sanger sequencing of a PCR fragment from the PEST domain (exon 34, chr9:139,390,619–139,391,290), while SF3B1 (exons 13–16) and TP53 (exons 2–11) were analyzed by DHPLC (WAVE® 3500HT, Transgenomic Inc.) followed by sequencing of aberrant fragments.
NOTCH1mut were found in 13/97 (13.4%), SF3B1mut in 17/97 (17.5%), and TP53mut in 37/99 (37.4%) cases. Two different types of NOTCH1mut (c.7541_7542delCT, n=11; c.7444delC, n=2) and 13 different point mutations of SF3B1 were identified. All SF3B1mut were heterozygous missense mutations resulting into 11 different amino acid changes in exons 14, 15 and 16. Of note, in none of the 97 cases concurrent NOTCH1mut and SF3B1mut was found. IGHV was unmutated in 71/90 (79%) cases overall, in 100% of NOTCH1mut (p=.062) and in 60.0% of SF3B1mut (p=.078). Moreover, SF3B1mut was associated with 11q deletion (p=.042), but we observed no significant correlation of NOTCH1mut with trisomy 12 (p=.321).
TP53mut was found together with NOTCH1mut in 3/12 (25%, p=.349) and with SF3B1mut in 7/17 (41%, p=1.000) cases.
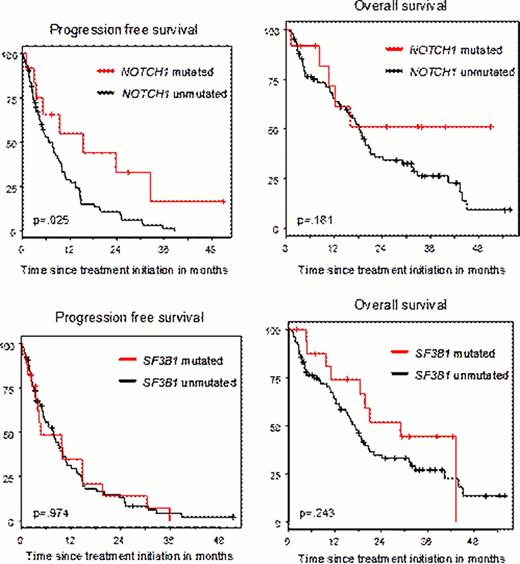
The overall response rates (ORR) among the 97 cases was 34% (3% CR and 31% PR). After a median follow-up time of 33.9 months, the median progression-free survival (PFS) was 7.7 months, and the median overall survival (OS) was 18.3 months. The ORR was not significantly different when comparing the groups with or without mutations (50.0% vs. 34.2% for NOTCH1, p=.341 and 41.2% vs. 35.1 % for SF3B1, p=.781, respectively). PFS in NOTCH1mut cases was significantly superior as compared to wild type cases (median 15.47 months vs. 6.74 months, p=.025, see Figure) but there was no significant difference in OS (median not reached vs. 18.3 months, p=.181). For SF3B1mut, we found no significant difference between mutated and wild type cases in PFS (median 4.76 vs. 7.72, p=.974) and OS (median 29.0 vs. 17.1, p=.243). Multivariate analysis of PFS and OS was performed including age, ECOG performance status, WBC, thymidine kinase, β2-microglobulin, IGHV status, 11q deletion, 17p deletion, NOTCH1mut, SF3B1mut and TP53mut as potential factors. Regarding PFS, ECOG performance status > 1 (HR 2.080, p=.057), increased thymidine kinase (cont.) (HR 1.479, p=.007) and elevated β2-microglobulin (cont.) (HR 1.188, p=.0003) emerged as independent unfavorable prognostic factors, whereas NOTCH1mut (HR 0.375, p=.049) was a favorable marker. Regarding OS, none of the gene mutations showed significant prognostic impact, whereas age at increments of 10 years (HR 1.035, p=.024), ECOG performance status > 1 (HR 2.575, p=.017) and increased thymidine kinase (HR 1.568, p=.002) were identified as independent unfavorable factors.
In summary, our analysis performed in a multicenter trial of fludarabine-refractory CLL patients showed NOTCH1mut in 13.4%, SF3B1mut in 17.5% and TP53mut in 37.4% cases, with NOTCH1mut and SF3B1mut being mutually exclusive. As compared to non-refractory CLL cohorts, the incidence of NOTCH1mut and SF3B1mut do not appear to be increased which is in contrast to TP53mut. NOTCH1mut was found as a favorable prognostic marker for PFS, but none of the mutations impacted OS in this trial.
Hallek:Roche: Consultancy, Honoraria, Research Funding. Stilgenbauer:Genzyme: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract