Abstract
Immunotherapy with therapeutic idiotype vaccines offers promise for treatment of B-cell malignancies. However, identification of novel immunogenic lymphoma-associated antigens that are universally expressed is necessary to overcome the barriers of patient-specific idiotype vaccines. Here, we determined whether T-cell leukemia/lymphoma 1 (TCL1) oncoprotein encoded by the TCL1 gene could be a target for immunotherapy of B-cell malignancies. We show that TCL1 mRNA and protein are selectively expressed in normal B cells but markedly hyperexpressed in multiple human B-cell lymphomas, including follicular lymphoma, chronic lymphocytic leukemia, mantle cell lymphoma, diffuse large B-cell lymphoma, and splenic marginal zone B-cell lymphoma. We demonstrated that TCL1-specific CD8+ T cells can be generated from HLA-A*0201 (HLA-A2)+ normal donors and identified TCL171-78 (LLPIMWQL) as the minimal epitope recognized by these T cells. More importantly, TCL171-78 peptide-specific T cells were present in the peripheral blood and tumor-infiltrating lymphocytes of lymphoma patients, could be expanded in vitro, and lysed autologous tumor cells but not normal B cells in an HLA-A2–restricted manner. Our results suggest that TCL1 is naturally processed and presented on the surface of lymphoma cells for recognition by cytotoxic T cells and can serve as a novel target for development of immunotherapeutic strategies against common B-cell lymphomas.
Introduction
Malignancies of B-cell origin are highly responsive to combination chemotherapy, and complete remissions can be induced in most patients. The use of rituximab, an anti-CD20 monoclonal antibody, in combination with chemotherapy has improved the overall and complete response rates, progression-free survival, overall survival, and curability of patients with B-cell non-Hodgkin lymphomas.1–3 However, relapse remains a significant cause of treatment failure, and novel treatments are needed to eradicate minimal residual disease to further improve clinical outcome in these patients.
Therapeutic agents used to eradicate minimal residual disease should ideally be directed at different targets and have different mechanisms of action than agents used in induction therapy. They also should be safe with minimal adverse effects because they may need to be administered as maintenance therapy over several months. Therapeutic vaccines have many of these desirable features because they can target different antigens on the lymphoma tumor cells than those targeted by rituximab or chemotherapy agents, and they are safe and well tolerated.4 Furthermore, by inducing immunologic memory and polyclonal humoral and cellular immune responses, vaccines may potentially produce a sustained antitumor effect, and unlike monoclonal antibodies, they may prevent the emergence of antigen-loss variants. Thus, therapeutic vaccines against lymphomas can be complementary to passive immunotherapeutic agents such as monoclonal antibodies and cytotoxic chemotherapeutic agents and could be ideal for eradicating minimal residual disease.
Several groups have used the clonal tumor immunoglobulin expressed on the surface of mature B-cell malignancies, termed idiotype, as a tumor-specific antigen for development of therapeutic vaccines against lymphomas.5–11 Idiotype vaccines were shown to be safe and induced sustained antitumor antibody and CD4+ and CD8+ T-cell responses in patients with follicular lymphoma (FL), chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), and diffuse large B-cell lymphoma (DLBCL).5–11 Furthermore, idiotype vaccines induced molecular remissions when administered after standard chemotherapy.10 A recently completed randomized, double blind, multicenter phase 3 clinical trial showed that idiotype vaccination improves disease-free survival when administered in the setting of minimal residual disease in FL, providing proof of principle that therapeutic vaccines can improve clinical outcome in these patients.12 However, a major limitation of idiotype vaccines is the requirement for a custom-made product for each patient that makes the manufacturing of the vaccine expensive, laborious, and time-consuming. To overcome these difficulties, identification of novel lymphoma-associated antigens that are shared between patients and universally expressed in multiple B-cell malignancies is necessary.
To prevent the induction of autoimmunity, tumor-associated antigens should be uniquely expressed or hyperexpressed in tumors compared with normal tissues. The T-cell leukemia/lymphoma 1 (TCL1) oncoprotein encoded by the TCL1 gene (also called TCL1A) was shown to be aberrantly expressed in many B-cell malignancies.13–16 Therefore, we evaluated whether it can serve as a shared lymphoma-associated antigen. TCL1 is primarily expressed during early embryogenesis in fetal lymphoid tissues, including liver, yolk sac, thymus, spleen, tonsil, and bone marrow.17 In adults, TCL1 expression is restricted to germ cells in the testis and precursor B and T lymphocytes. During B-cell development, TCL1 expression begins at the pro-B cell stage and is present until they differentiate into naive B cells. After antigen activation, TCL1 is down-regulated in the germinal center (GC) B cells and is silenced in post-GC memory B cells and plasma cells.15 During T-cell development, TCL1 is expressed in CD3−CD4−CD8− thymocytes but is absent in mature T cells in the periphery.18,19 TCL1 was demonstrated to function as a coactivator of Akt (also called protein kinase B) and thereby promote cell proliferation and survival.20 The tumorigenic role for TCL1 was established in transgenic mice where ectopic expression of human TCL1 in B cells resulted in the development of B-cell malignancies.21–23 Furthermore, TCL1 expression was associated with adverse clinical outcome in CLL, MCL, and DLBCL.13,24–26 Thus, the aberrant expression in multiple B-cell malignancies, the restricted expression in normal tissues, and possible causative role in B-cell lymphomas suggested that TCL1 might serve as a shared tumor-associated antigen in lymphomas.
Here, we characterized the expression of TCL1 in B-cell lymphomas and normal tissues, identified TCL171-78 as the minimal epitope that binds to HLA-A*0201 (HLA-A2), and we show that cytotoxic T lymphocytes (CTLs) specific to this peptide can efficiently kill lymphoma cell lines and primary human lymphoma cells. Our results suggest that TCL1 is naturally processed and presented on the surface of primary lymphoma cells for recognition by CTL in an HLA-restricted manner and can potentially serve as a shared lymphoma-associated antigen for therapeutic vaccine development against multiple B-cell malignancies.
Methods
Patient samples
The Institutional Review Board of The University of Texas MD Anderson Cancer Center approved the study. An informed consent was obtained in accordance with the Declaration of Helsinki before collection of patient samples. Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples by density gradient separation. Tissue samples were processed into single-cell suspension and cryopreserved in aliquots.
Cell lines
MCL cell lines Mino and Jeko-1, Burkitt lymphoma cell line Daudi, erythroleukemia cell line K562, breast cancer cell line MCF-7, and T2 hybridoma cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 10mM HEPES, 1 × GlutaMAX, 50μM β-mercaptoethanol, 1mM sodium pyruvate, 100 U/mL penicillin + 100 μg/mL streptomycin, and 10 μg/mL gentamicin (all from Invitrogen) at 37°C and 5% CO2 in air.
Tumor and immune cell subset isolation
Tumor cells, T cells from tumor samples, and normal peripheral blood B and T cells were isolated by magnetic cell separation (Miltenyi Biotec), and the purity was confirmed by flow cytometry as described previously.27 The procedure yielded > 90% purity of tumor B cells, tumor infiltrating T cells, and normal B and T cells (data not shown).
Reagents
Mouse anti–human antibodies against CD3, CD4, CD8, CD10, CD20, interferon (IFN)–γ, CD38, IgD, CD19, CD34, CD16, CD56, PD-1, CD160, CTLA-4, and CD244 were obtained from BD Biosciences; anti–human BTLA was from Biolegend; and anti–human LAG3 was from Alexis Biochemicals. Enzyme-linked immunosorbent assay (ELISA) kits for IFN-γ were obtained from R&D Systems. All peptides were synthesized by Sigma-Aldrich to > 70% purity and dissolved in dimethyl sulfoxide (Sigma-Aldrich). Phycoerythrin (PE)–conjugated tetramers were synthesized by MHC Tetramer Lab, Protein Chemistry Core of Baylor College of Medicine (Houston, TX). TCL1 and β-actin antibodies for Western blotting were obtained from Cell Signaling Technology. Anti–human HLA-ABC (clone W6/32; eBioscience), anti–HLA-DPDQDR (clone TÜ39; BD Biosciences), anti-HLA A2 (clone BB7.2; BD Biosciences), and mouse IgG2a isotype control (eBioscience) were used for HLA-blocking studies. Blocking antibody against HLA-B and -C was a kind gift from Paul Robbins (National Cancer Institute, National Institutes of Health, Bethesda, MD).
Analysis of cDNA microarray data
The relative expression of TCL1 mRNA in lymphoma cells and normal tissues was determined from publicly available cDNA microarray datasets from the Oncomine database.28 Log2-transformed data were analyzed for relative levels of gene expression.
Real-time PCR
FirstChoice Human Total RNA Survey Panel containing 20 normal human tissues total RNA and human lymph node total RNA was obtained from Applied Biosystems. Total RNA was extracted from purified lymphoma cells or peripheral blood B and T cells using TRIzol (Invitrogen). Approximately 10 μg of total RNA from each source was reverse transcribed into cDNA with SuperScript III kit (Invitrogen). Quantitative PCR was performed with TaqMan real-time PCR kit (Applied Biosystems) and TCL1 (Applied Biosystems, catalog no. Hs00951350_m1) and β-actin primers (Applied Biosystems, catalog no. Hs99999903_m1) using the following conditions: 50°C for 2 minutes, 94°C for 10 minutes, followed by 94°C for 15 seconds, 60°C for 60 seconds for 40 cycles on a StepOne/StepOnePlus real-time PCR system (Applied Biosystems). The expression of TCL1 mRNA relative to the β-actin mRNA was calculated in each sample.
Immunohistochemistry
All tumor tissues were obtained from the MD Anderson Lymphoma Tissue Bank. In brief, formalin-fixed, paraffin-embedded tissue sections from different lymphomas (DLBCL, FL, MCL, CLL, and splenic marginal zone lymphoma [SMZL]) and reactive tonsils were deparaffinized, and heat-induced antigen retrieval was performed as per the manufacturer's instructions (Vector Laboratories). Sections were incubated for 10 minutes with TBS containing 3% bovine serum albumin (BSA) to block nonspecific staining. Next, specimens were incubated overnight at 4°C with the primary antibody against TCL-1 (Cell Signaling Technology). Antibody binding was detected using a secondary antibody in blocking buffer and staining was visualized using 3,3′-diaminobenzidine (Vector Laboratories). Digital photomicrographs were acquired using DP Controller Version 3.31 software mounted on a BX41 inverted microscope (Olympus).
Flow cytometry
For intracellular staining, cells were first surface stained with CD3 and CD20 and then fixed and permeabilized using Cytofix/Cytoperm Plus Fixation/Permeabilization kit (BD Biosciences) as per the manufacturer's instructions. Cells were then stained with mouse anti–human-TCL1 antibody (eBioscience) or Alexa Fluor 647 Mouse IgG2b isotype control antibody (eBiosciences) for 30 minutes at 4°C. After 2 washes, samples were acquired on an FACSCalibur flow cytometer (BD Biosciences) and analyzed using CellQuest Pro Version 5.1 (BD Biosciences) or FlowJo Version 7.6.5 software (TreeStar). Intracellular cytokine staining was performed as described previously.27 For tetramer staining, PE-conjugated TCL171-78 tetramer or HIV Gag77-85 tetramer and FITC-conjugated mouse anti–human CD8 antibody were mixed with the cells in a 50-μL volume for 30 minutes at room temperature, washed twice, and analyzed by a flow cytometry.
Western blotting
Approximately 50 μg of total cell protein was extracted from tumor or normal B cells after cell lysis in a buffer composed of 50mM Tris-Cl (pH 7.4), 5mM EDTA, 150mM NaCl, 0.5% Triton-X 100, 1 mg/mL leupeptin and aprotinin, and 1mM PMSF. Protein content of the lysates was quantified by the Bradford assay (Bio-Rad Laboratories), and 25 μg of total protein was dissolved in Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer before separation in 15% SDS-PAGE gels. After electrophoresis, proteins were transferred to a nitrocellulose membrane (Bio-Rad Laboratories) for Western blotting with mouse anti–human-TCL1 antibody (1:200 dilution; eBioscience) for 1 hour at room temperature or at 4°C overnight. Bound antibodies were detected with goat anti–mouse horseradish peroxidase–conjugated secondary antibody diluted at 1:10 000 and SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
HLA-A2-binding assay
Transporter associated with Antigen Processing (TAP)–deficient T2 cells were incubated with 20 μg/mL peptides and 3 μg/mL β2-microglobulin (Sigma-Aldrich) for 24 hours at 37°C and 5% CO2 in air. The cells were then stained with anti–HLA-A2 PE antibody (BD Biosciences), and the level of HLA-A2 expression was determined by flow cytometry. The binding affinity of the peptide to HLA-A2 (T2 binding index) was quantified as follows: (mean fluorescence intensity [MFI] of HLA-A2 with peptide − MFI of HLA-A2 without peptide)/(MFI of HLA-A2 without peptide).
Generation of peptide-specific T-cell lines
Peptide-specific T-cell lines were generated from HLA-A2+ normal donors and patients using methods reported previously.29 In brief, PBMCs (1 × 105 cells/well) were stimulated with 10 μg/mL of each peptide in quadruplicate in a 96-well U-bottomed microculture plate (Corning Life Sciences) in 200 μL of culture medium. The culture medium consisted of 50% AIM-V medium (Invitrogen), 50% RPMI 1640 medium (Invitrogen), 10% human AB serum (Valley Biomedical), and 100 IU/mL of interleukin (IL)–2. Cells were restimulated with the corresponding peptide every 3 days. After 5 stimulations, T cells from each well were washed and incubated with T2 cells in the presence or absence of the corresponding peptide. After 18 hours, the production of IFN-γ was determined in the supernatants by ELISA. T cells that secreted large amount of IFN-γ were further expanded by rapid expansion protocol to obtain sufficient number of cells for additional functional assays as described previously.27
Cytotoxicity assay
T2 cells were cultured overnight with 40 μg/mL peptide in the presence of 3 μg/mL β2-microglobulin, washed twice, labeled with chromium-51 (51Cr) for 1 hour at 37°C, and used as targets in cytotoxicity assay. Primary tumor cells were isolated from PBMCs or biopsy samples of lymphoma patients by magnetic cell separation before labeling with 51Cr. Target cells (1 × 104 cells/well) were incubated with the effector T cells at the indicated ratios in 96-well round-bottomed plates at 37°C for 4 hours, and target cell lysis was determined by chromium release into the supernatants. Spontaneous lysis was determined from the supernatant of target cells cultured without effector cells, and the maximal lysis was determined from the supernatant of target cells cultured with 1% Triton X-100. The specific lysis of target cells was calculated as follows: (experimental lysis − spontaneous lysis) × 100/(maximal lysis − spontaneous lysis). For HLA-blocking studies, target cells were incubated for 30 minutes with 20 μg/mL of the anti-HLA blocking antibodies before coculturing with the effector T cells. All assays were performed in triplicate wells and repeated at least 2 times.
Statistical analysis
The Student t test was used to compare various experimental groups. P values < .05 were considered statistically significant. Unless otherwise indicated, means and standard deviations are shown.
Results
TCL1 is hyperexpressed in multiple B-cell lymphomas
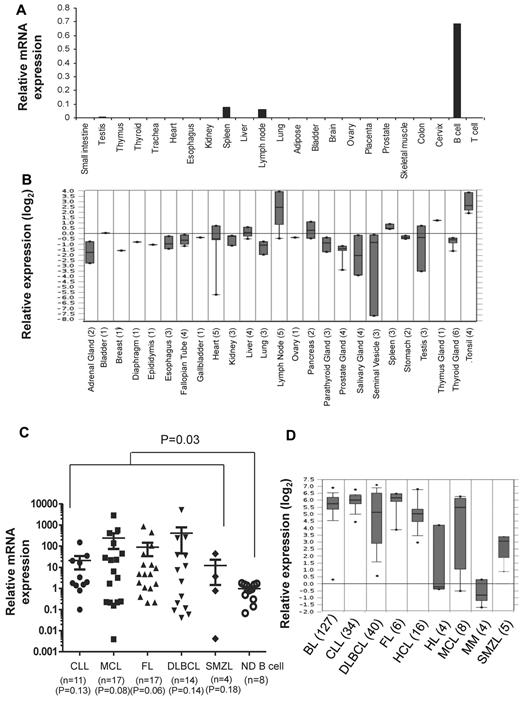
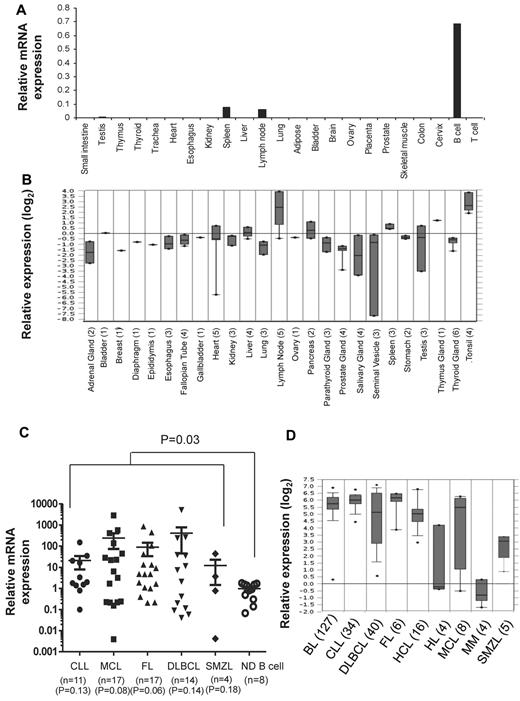
To determine the expression pattern of TCL1 in normal adult tissues, we performed real-time PCR assay on RNA derived from 21 tissues and peripheral blood B and T cells obtained from normal adults. Consistent with previous reports,13,15,16,30 we detected TCL1 mRNA in peripheral blood B cells and 2 normal lymphoid tissues, lymph node and spleen (Figure 1A). We also found very low levels of TCL1 mRNA in testis. However, TCL1 mRNA was not detected in peripheral blood T cells and tissues from small intestine, thymus, thyroid, trachea, heart, esophagus, kidney, liver, lung, adipose tissue, bladder, brain, ovary, placenta, prostate, skeletal muscle, colon, and cervix. Our results were also consistent with the expression pattern of TCL1 mRNA observed in cDNA microarray data from normal tissues in Oncomine database (Figure 1B). Because TCL1 is selectively expressed in normal B cells (Figure 1A), we compared the expression level of TCL1 mRNA in B-cell lymphomas with peripheral blood B cells from normal donors. We observed that TCL1 mRNA was significantly hyperexpressed in tumor cells when all lymphomas were compared with normal B cells (P = .03; Figure 1C). However, when each lymphoma subtype was compared with normal B cells, we found that there was a trend toward higher expression of TCL1 in lymphoma samples, but it did not reach statistical significance, probably because of the smaller sample size. Analysis of publicly available cDNA microarray data similarly showed that TCL1 mRNA was hyperexpressed in FL, CLL, MCL, SMZL, and DLBCL (Figure 1D). In addition, TCL1 mRNA was hyperexpressed in Burkitt lymphoma and hairy cell leukemia but not in Hodgkin lymphoma and multiple myeloma (Figure 1D).
TCL1 mRNA is hyperexpressed in multiple B-cell lymphomas. (A,C) Total RNA was extracted from normal human tissues, normal human peripheral blood B and T cells, and from primary human lymphoma cells; reverse transcribed into cDNA; and then quantitative PCR was performed for TCL1 and β-actin. (A) Expression of TCL1 mRNA relative to β-actin is shown in normal human tissues. (C) Expression of TCL1 mRNA in primary human lymphoma cells is shown relative to TCL1 expression in normal human peripheral blood B cells. Horizontal lines represent the median value for each group. P value on the top of graph represents comparison between normal donor B cells and all lymphomas. P values below the graph represent comparison between normal donor B cells and each lymphoma subtype. All P values were calculated by 2-tailed Student t test. (B,D) Expression of TCL1 mRNA in normal human tissues (B) and different human lymphoma subtypes (D) was determined from publicly available cDNA microarray datasets from the Oncomine database. The number of samples for each tissue type is shown in brackets. CLL indicates chronic lymphocytic leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; SMZL, splenic marginal zone B-cell lymphoma; HL, Hodgkin lymphoma; BL, Burkitt lymphoma; MM, multiple myeloma; and HCL, hairy cell leukemia.
TCL1 mRNA is hyperexpressed in multiple B-cell lymphomas. (A,C) Total RNA was extracted from normal human tissues, normal human peripheral blood B and T cells, and from primary human lymphoma cells; reverse transcribed into cDNA; and then quantitative PCR was performed for TCL1 and β-actin. (A) Expression of TCL1 mRNA relative to β-actin is shown in normal human tissues. (C) Expression of TCL1 mRNA in primary human lymphoma cells is shown relative to TCL1 expression in normal human peripheral blood B cells. Horizontal lines represent the median value for each group. P value on the top of graph represents comparison between normal donor B cells and all lymphomas. P values below the graph represent comparison between normal donor B cells and each lymphoma subtype. All P values were calculated by 2-tailed Student t test. (B,D) Expression of TCL1 mRNA in normal human tissues (B) and different human lymphoma subtypes (D) was determined from publicly available cDNA microarray datasets from the Oncomine database. The number of samples for each tissue type is shown in brackets. CLL indicates chronic lymphocytic leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; SMZL, splenic marginal zone B-cell lymphoma; HL, Hodgkin lymphoma; BL, Burkitt lymphoma; MM, multiple myeloma; and HCL, hairy cell leukemia.
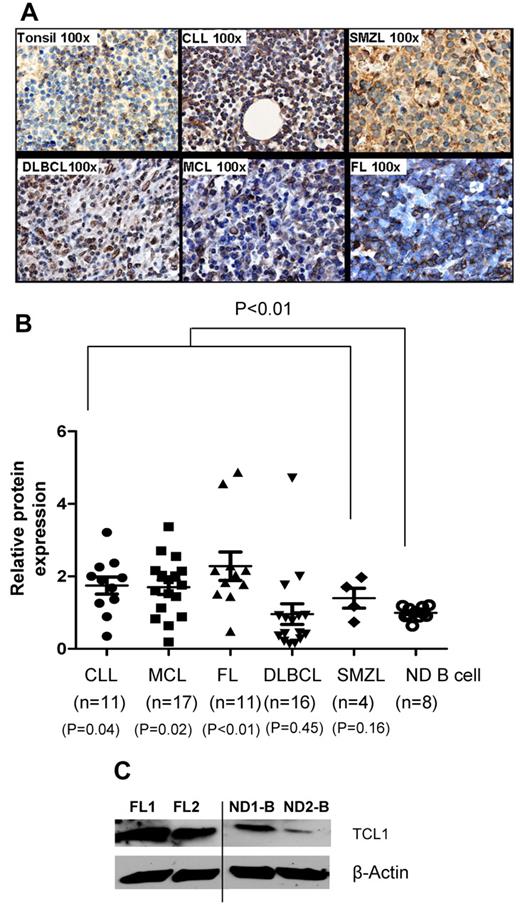
To determine the expression of TCL1 protein in lymphomas, we performed immunohistochemistry, flow cytometry, and Western blotting. Compared with reactive tonsils or peripheral blood B cells, TCL1 protein was hyperexpressed in FL, CLL, MCL, DLCL, and SMZL (Figure 2A-C). By flow cytometry, we found that the TCL1 protein was significantly hyperexpressed in tumor cells when all lymphomas were compared with normal B cells (P < .01) but only in CLL (P = .04), MCL (P = .02), and FL (P < .01) when they were compared individually (Figure 2B). Furthermore, we found that TCL1 was not expressed in CD34+ hematopoietic stem cells but is expressed in a subset of pro-B, pre-B, and immature B cells in normal human bone marrow samples (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To determine the subset of mature B cells that express TCL1, we analyzed normal human tonsil samples. We found that TCL1 was present in naive and GC B cells and was down-regulated in memory B cells and plasma cells (supplemental Figure 1B). Among immature thymocytes, TCL1 was expressed in a subset of double-negative thymocytes but was very low or absent in double-positive, CD4 single-positive, and CD8 single-positive thymocytes (supplemental Figure 1C). This pattern of expression of TCL1 in normal B cell progenitors, mature B cell subsets, and immature thymocytes was consistent with prior reports in the literature.13–16,30 Taken together, our results suggest that in adults, TCL1 is expressed at low levels in mature B cells and their precursors and is hyperexpressed in multiple B-cell lymphomas and therefore may potentially serve as a shared tumor-associated antigen for development of active immunotherapy against these malignancies.
TCL1 protein is hyperexpressed in multiple B-cell lymphomas. (A) Expression of TCL1 protein was determined by immunohistochemistry in formalin-fixed, paraffin-embedded primary human lymphoma tissues (FL, n = 5; CLL, n = 2; MCL, n = 3; DLBCL, n = 2; and SMZL, n = 3) and reactive human tonsils (n = 4). Images shown are the original magnification, ×100. (B) Intracellular staining for TCL1 was performed on primary human lymphoma cells and normal human peripheral blood B cells, and samples were analyzed by flow cytometry. The relative expression of TCL1 was calculated as follows: mean fluorescence intensity of TCL1 in test sample/mean fluorescence intensity of TCL1 in normal B cells. Horizontal lines represent the median value for each group. P value on the top of graph represents comparison between normal donor B cells and all lymphomas. P values below the graph represent comparison between normal donor B cells and each lymphoma subtype. All P values were calculated by 2-tailed Student t test (C) TCL1 protein expression was determined by Western blotting in 2 FL and 2 normal donor (ND) peripheral blood B cells. β-Actin protein expression was used as loading control. Vertical line inserted between lanes 2 and 3 to indicate repositioned gel lanes obtained from the same gel.
TCL1 protein is hyperexpressed in multiple B-cell lymphomas. (A) Expression of TCL1 protein was determined by immunohistochemistry in formalin-fixed, paraffin-embedded primary human lymphoma tissues (FL, n = 5; CLL, n = 2; MCL, n = 3; DLBCL, n = 2; and SMZL, n = 3) and reactive human tonsils (n = 4). Images shown are the original magnification, ×100. (B) Intracellular staining for TCL1 was performed on primary human lymphoma cells and normal human peripheral blood B cells, and samples were analyzed by flow cytometry. The relative expression of TCL1 was calculated as follows: mean fluorescence intensity of TCL1 in test sample/mean fluorescence intensity of TCL1 in normal B cells. Horizontal lines represent the median value for each group. P value on the top of graph represents comparison between normal donor B cells and all lymphomas. P values below the graph represent comparison between normal donor B cells and each lymphoma subtype. All P values were calculated by 2-tailed Student t test (C) TCL1 protein expression was determined by Western blotting in 2 FL and 2 normal donor (ND) peripheral blood B cells. β-Actin protein expression was used as loading control. Vertical line inserted between lanes 2 and 3 to indicate repositioned gel lanes obtained from the same gel.
TCL1-specific T cells can be generated from normal donors
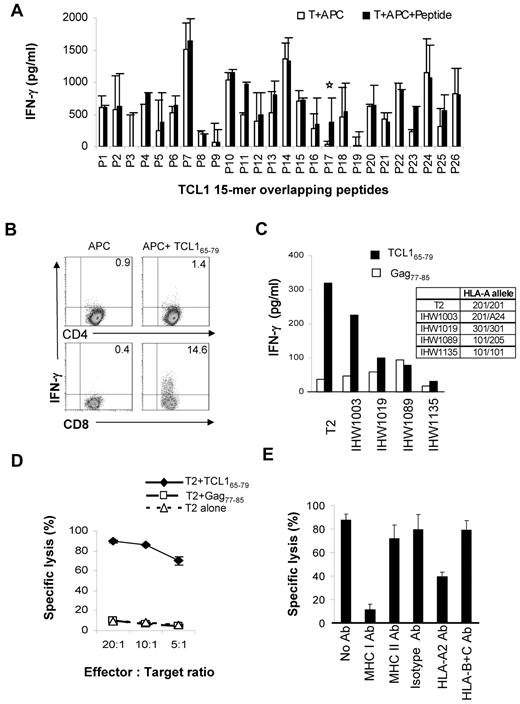
To determine whether TCL1 is immunogenic, we synthesized overlapping 15-mer peptides spanning the entire length of the TCL1 protein (supplemental Table 1) and stimulated PBMC from HLA-A2+ normal donors to generate TCL1-specific T cells, as described under “Methods.”29 We found that TCL165-79 peptide (TQIGPSLLPIMWQLY) consistently induced peptide-specific IFN-γ–producing T cells in 3 HLA-A2+ normal donors (Figure 3A). By intracellular cytokine assay, we observed that CD8+ but not CD4+ T cells produced IFN-γ in response to TCL165-79 peptide stimulation (Figure 3B and supplemental Figure 2).
TCL1-specific cytotoxic T cells can be generated from normal donors. (A) PBMCs from HLA-A2+ normal donors were stimulated with overlapping 15-mer peptides spanning the entire length of the TCL1 protein (supplemental Table 1). T cells from each condition were washed and incubated with autologous CD3-depleted PBMCs as antigen-presenting cells (APC) in the presence or absence of the corresponding peptide. After 18 hours, the production of IFN-γ was determined in the supernatants by ELISA. Representative data from one of 3 normal donors tested is shown. (B) T-cell lines generated from HLA-A2+ normal donors using TCL165-79 peptide were incubated with APC in the presence or absence of the TCL165-79 peptide or control HIV Gag77-85 peptide, and intracellular cytokine assay was performed. The percentage of CD4+ and CD8+ T cells producing IFN-γ are shown. (C) TCL165-79 peptide-specific T cells were incubated with T2 cells or EBV-transformed B lymphoblastoid cell lines (IHW1003, IHW1019, IHW1089, and IHW1135) mismatched at their MHC class I locus in the presence of the TCL165-79 peptide or control HIV Gag77-85 peptide. IFN-γ production was determined as in panel A. The HLA-A alleles for each of the cell lines are shown. (D,E) A 4-hour 51Cr-release cytotoxicity assay was performed. TCL165-79 peptide-specific T cells were incubated with T2 cells alone or T2 cells pulsed with TCL165-79 peptide or control HIV Gag77-85 peptide, at various effector:target ratios. For MHC blocking assay, an effector: target ratio of 20:1 was used in the presence or absence of isotype control antibody or blocking antibodies (10 μg/mL for each antibody) against MHC class I, MHC class II, HLA-A2, or HLA-B and -C. All cytotoxicity assays were performed in triplicate wells, and mean and standard deviation are shown.
TCL1-specific cytotoxic T cells can be generated from normal donors. (A) PBMCs from HLA-A2+ normal donors were stimulated with overlapping 15-mer peptides spanning the entire length of the TCL1 protein (supplemental Table 1). T cells from each condition were washed and incubated with autologous CD3-depleted PBMCs as antigen-presenting cells (APC) in the presence or absence of the corresponding peptide. After 18 hours, the production of IFN-γ was determined in the supernatants by ELISA. Representative data from one of 3 normal donors tested is shown. (B) T-cell lines generated from HLA-A2+ normal donors using TCL165-79 peptide were incubated with APC in the presence or absence of the TCL165-79 peptide or control HIV Gag77-85 peptide, and intracellular cytokine assay was performed. The percentage of CD4+ and CD8+ T cells producing IFN-γ are shown. (C) TCL165-79 peptide-specific T cells were incubated with T2 cells or EBV-transformed B lymphoblastoid cell lines (IHW1003, IHW1019, IHW1089, and IHW1135) mismatched at their MHC class I locus in the presence of the TCL165-79 peptide or control HIV Gag77-85 peptide. IFN-γ production was determined as in panel A. The HLA-A alleles for each of the cell lines are shown. (D,E) A 4-hour 51Cr-release cytotoxicity assay was performed. TCL165-79 peptide-specific T cells were incubated with T2 cells alone or T2 cells pulsed with TCL165-79 peptide or control HIV Gag77-85 peptide, at various effector:target ratios. For MHC blocking assay, an effector: target ratio of 20:1 was used in the presence or absence of isotype control antibody or blocking antibodies (10 μg/mL for each antibody) against MHC class I, MHC class II, HLA-A2, or HLA-B and -C. All cytotoxicity assays were performed in triplicate wells, and mean and standard deviation are shown.
To determine whether HLA-A2 is the restriction element for the TCL165-79 peptide-reactive CD8+ T cells, we cocultured the T cells with a panel of peptide-pulsed EBV-transformed B lymphoblastoid cell lines that were mismatched at the HLA-A locus. We observed significant production of IFN-γ when the TCL165-79 peptide was presented by IHW1003 cells that expressed HLA-A2 and HLA-A24 but not by antigen-presenting cells expressing other HLA-A alleles (Figure 3C). Interestingly, T2 cells also induced IFN-γ production by TCL165-79 peptide-reactive T cells. Because T2 cells are TAP deficient, it is unlikely that the TCL165-79 peptide was processed and presented as a shorter peptide by T2 cells. Because peptides longer than 9 amino acids also may bind to MHC class I molecules,31–35 we reasoned that the unprocessed TCL165-79 peptide was being presented by T2 cells. To further confirm this, we tested whether the TCL165-79 peptide-reactive T cells would lyse peptide-pulsed HLA-A2+ T2 cells. We observed significant lysis of TCL165-79 peptide-pulsed T2 cells but not unpulsed T2 cells or T2 cells pulsed with control HIV Gag77-85 peptide (Figure 3D). The lysis of TCL165-79 peptide-pulsed T2 cells was inhibited by MHC class I and HLA-A2 blocking antibodies but not by MHC class II and HLA-B and -C blocking antibodies or isotype control antibody, further confirming HLA-A*0201 as the restriction element for this peptide (Figure 3E). Collectively, these results suggested that the CD8+ T cells generated against TCL165-79 peptide were peptide-specific and recognized the peptide in the context of HLA-A2.
TCL171-78 is the minimal HLA-A2–binding epitope
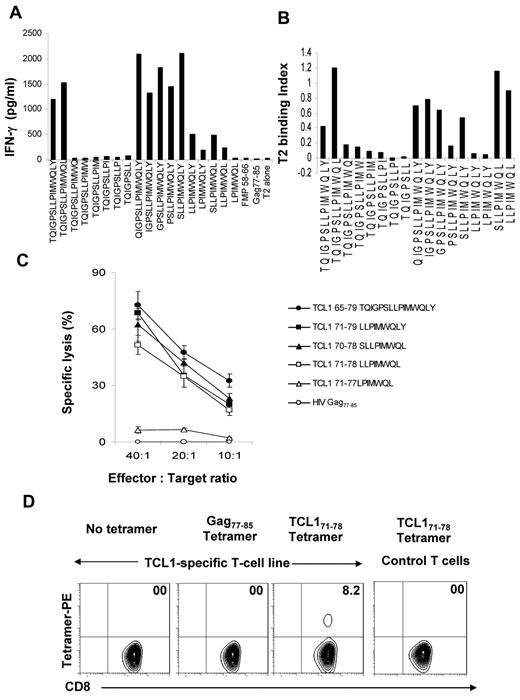
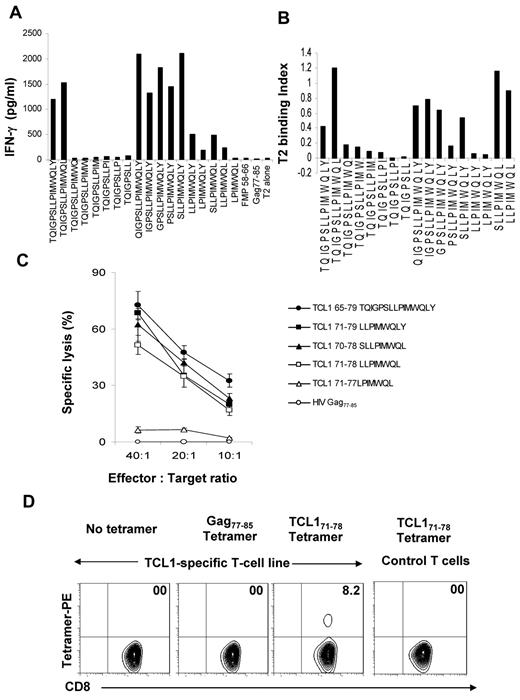
To determine the minimal immunogenic epitope in the TCL165-79 peptide, we generated a panel of peptides truncated progressively by one amino acid at the N and C termini and incubated the TCL165-79 peptide-specific T cells with T2 cells pulsed with each of the peptides. We found that TCL171-78 (LLPIMWQL) is the minimal epitope that induced significant IFN-γ production by the TCL165-79 peptide-specific T cells (Figure 4A). Analysis of the peptides in a standard functional peptide-binding assay using T2 cells demonstrated that the peptides that induced IFN-γ production by the TCL165-79 peptide-specific T cells bound to HLA-A2, including the TCL171-78 peptide (Figure 4B). Conversely, truncated peptides that did not show significant binding to HLA-A2 did not induce IFN-γ production. To further confirm that the TCL171-78 peptide is the minimal epitope recognized by the TCL165-79 peptide-specific T cells, we performed cytotoxicity assay. TCL165-79 peptide-specific T cells specifically lysed T2 cells pulsed with TCL170-79 (SLLPIMWQL), TCL171-79 (LLPIMWQLY), and TCL171-78 (LLPIMWQL) peptides but not TCL171-77 (LPIMWQL) peptide, indicating that TCL171-78 (LLPIMWQL) is the minimal epitope recognized by TCL165-79 peptide-specific T cells. Finally, we used a tetramer synthesized with the TCL171-78 peptide and found that 8.2% of the TCL165-79 peptide-specific CD8+ T cells stained tetramer positive (Figure 4D). By peptide titration experiments, we found that these T cells recognized peptide concentrations as low as 1nM, suggesting that they were of moderate avidity (supplemental Figure 3).36 Together, these results suggested that TCL171-78 peptide is the minimal HLA-A2–binding epitope recognized by CD8+ T cells.
TCL171-78 is the minimal HLA-A*0201–binding epitope. (A) TCL165-79 peptide-specific T cells were incubated in the presence or absence of T2 cells and TCL165-79–derived truncated peptides. Flu matrix peptide (FMP58-66) and Gag77-85 peptide were used as controls. After 18 hours, the production of IFN-γ was determined in the supernatants by ELISA. (B) T2 cells were incubated with TCL165-79–derived truncated peptides, and the HLA-A2 binding affinity of each peptide was determined by flow cytometry. The T2 binding index was calculated as described under “Methods.” (C) TCL165-79 peptide-specific T cells were incubated with T2 cells pulsed with TCL165-79–derived truncated peptides or control HIV Gag77-85 peptide, and a 4-hour 51Cr-release cytotoxicity assay was performed. The percentage of specific lysis is shown. (D) TCL165-79 peptide-specific CD8+ T cells or irrelevant T-cell line were stained with TCL171-78 or HIV Gag77-85 tetramers and analyzed by flow cytometry. The percentage of tetramer positive T cells is shown. Data in panels A to D are representative of 3 independent experiments each.
TCL171-78 is the minimal HLA-A*0201–binding epitope. (A) TCL165-79 peptide-specific T cells were incubated in the presence or absence of T2 cells and TCL165-79–derived truncated peptides. Flu matrix peptide (FMP58-66) and Gag77-85 peptide were used as controls. After 18 hours, the production of IFN-γ was determined in the supernatants by ELISA. (B) T2 cells were incubated with TCL165-79–derived truncated peptides, and the HLA-A2 binding affinity of each peptide was determined by flow cytometry. The T2 binding index was calculated as described under “Methods.” (C) TCL165-79 peptide-specific T cells were incubated with T2 cells pulsed with TCL165-79–derived truncated peptides or control HIV Gag77-85 peptide, and a 4-hour 51Cr-release cytotoxicity assay was performed. The percentage of specific lysis is shown. (D) TCL165-79 peptide-specific CD8+ T cells or irrelevant T-cell line were stained with TCL171-78 or HIV Gag77-85 tetramers and analyzed by flow cytometry. The percentage of tetramer positive T cells is shown. Data in panels A to D are representative of 3 independent experiments each.
TCL1-specific T cells induced lysis of primary human lymphoma cells
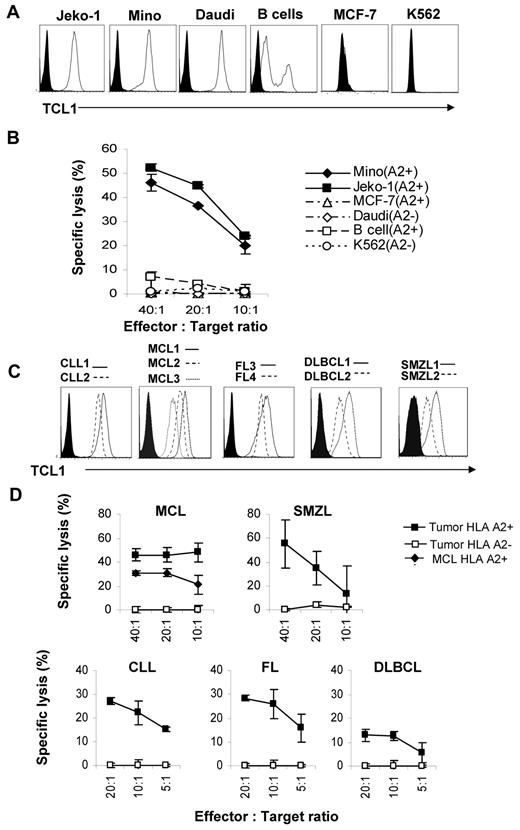
To determine whether TCL1 is processed and presented by tumor cells, we performed cytotoxicity assay against human lymphoma cell lines and primary lymphoma cells. Using flow cytometry, we observed that TCL1 was highly expressed in Mino, Jeko-1, and Daudi lymphoma cell lines but not in MCF-7 and K562 cell lines (Figure 5A). CD8+ T cells purified from TCL165-79 peptide-specific T cell lines (supplemental Figure 4A) induced significant lysis of HLA-A2+TCL1+ Mino and Jeko-1 cells but not HLA-A2−TCL1+ Daudi cells, HLA-A2−TCL1− K562 cells, or HLA-A2+TCL1− MCF-7 cells (Figure 5B). TCL1 was present at low levels in normal donor B cells and consistent with this, TCL165-79 peptide-specific T cells induced very low lysis of HLA-A2+ normal donor B cells. When various normal B cell subsets were used as targets, we found that TCL1-specific T cells had low cytotoxic activity against naive and GC B cells but not memory B cells (supplemental Figure 4B). Furthermore, consistent with the expression pattern of TCL1 (supplemental Figure 1A), TCL1-specific T cells had no cytotoxic activity against HLA-A2+ CD34+ hematopoietic stem cells (supplemental Figure 4B). We also observed that the TCL165-79 peptide-specific T cells induced significant lysis of HLA-A2+ TCL1+ primary tumor cells obtained from patients with FL, CLL, MCL, DLBCL, and SMZL but not HLA-A2−TCL1+ tumor cells (Figure 5C-D). These results suggested that TCL1 is naturally processed and presented on the cell surface by the primary lymphoma cells for recognition by TCL1-specific CD8+ T cells in an HLA-A2–restricted manner.
TCL165-79 peptide-specific T cells lysed human lymphoma cell lines and primary lymphoma cells. (A,C) Intracellular staining for TCL1 was performed on tumor cell lines and normal donor peripheral blood B cells (A) and primary human lymphoma cells (C). Black histograms represent cells stained with isotype control antibody, and open histograms represent staining with TCL1 antibody. (B,D) CD8+ T cells purified from TCL165-79 peptide-specific T cell lines by magnetic cell separation were incubated with tumor cell lines and normal peripheral blood B cells (B) or primary human lymphoma cells (D). A 4-hour 51Cr-release cytotoxicity assay was performed, and the percentage of specific lysis is shown. The HLA-A2 expression for each of the cell types is shown in the figures. Data in panels B and D are representative of 3 independent experiments.
TCL165-79 peptide-specific T cells lysed human lymphoma cell lines and primary lymphoma cells. (A,C) Intracellular staining for TCL1 was performed on tumor cell lines and normal donor peripheral blood B cells (A) and primary human lymphoma cells (C). Black histograms represent cells stained with isotype control antibody, and open histograms represent staining with TCL1 antibody. (B,D) CD8+ T cells purified from TCL165-79 peptide-specific T cell lines by magnetic cell separation were incubated with tumor cell lines and normal peripheral blood B cells (B) or primary human lymphoma cells (D). A 4-hour 51Cr-release cytotoxicity assay was performed, and the percentage of specific lysis is shown. The HLA-A2 expression for each of the cell types is shown in the figures. Data in panels B and D are representative of 3 independent experiments.
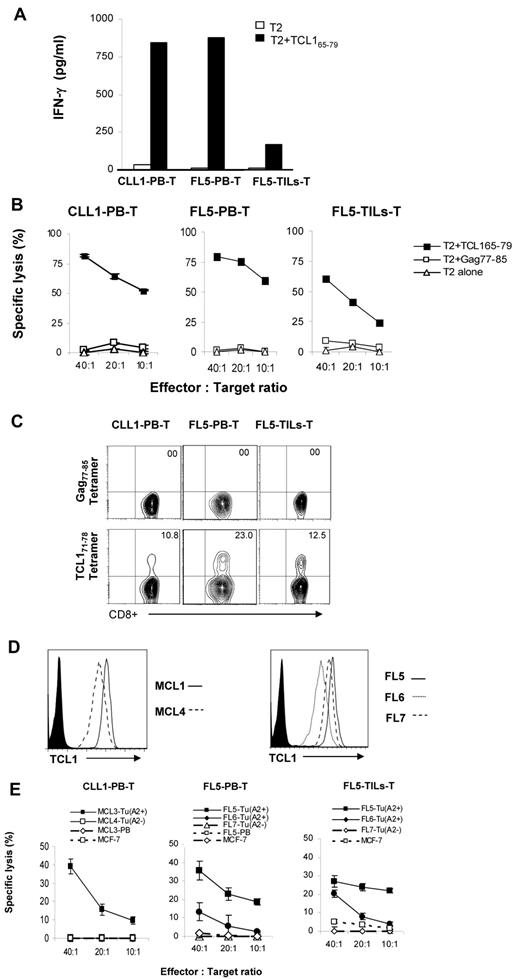
TCL171-78 peptide-specific T cells can be generated from lymphoma patients
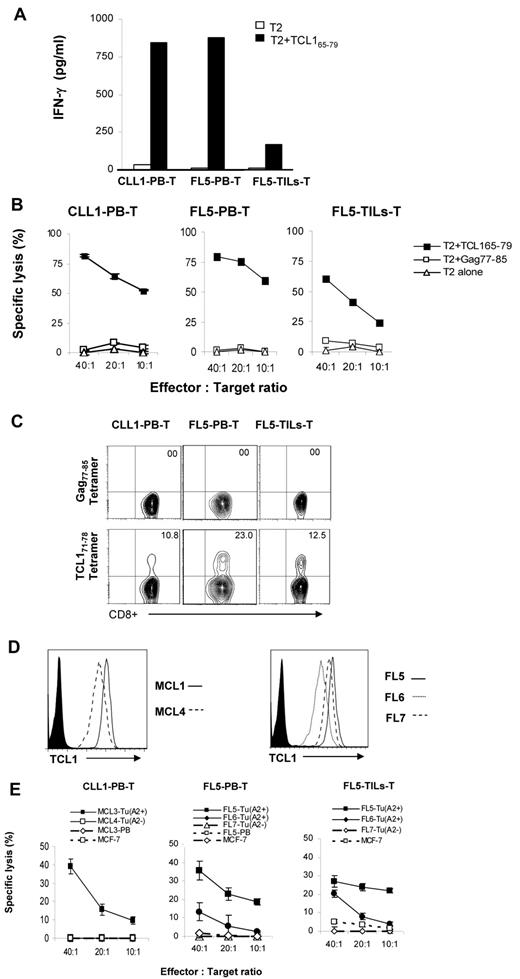
To determine whether TCL171-78 peptide-specific T cells can be generated from lymphoma patients, we stimulated PBMCs from a patient with CLL and PBMCs and intratumoral T cells from a patient with FL with TCL165-79 peptide. These patient-derived T-cell lines secreted significant amount of IFN-γ on incubation with TCL165-79 peptide-pulsed T2 cells (Figure 6A). Furthermore, these T cells lysed TCL165-79 peptide-pulsed T2 cells at high efficiency but not control peptide-pulsed or unpulsed T2 cells (Figure 6B). Tetramer staining using TCL171-78 peptide showed that 10.8% to 23% of the CD8+ T cells were tetramer positive (Figure 6C). To further confirm the cytotoxic activity of the patient-derived T-cell lines, we incubated the T cells with autologous or allogeneic primary lymphoma cells. We found that the patient-derived TCL165-79 peptide-specific T cells efficiently lysed HLA-A2+TCL1+ allogeneic or autologous primary lymphoma cells but not HLA-A2−TCL1+ lymphoma cells, HLA-A2+ tumor-free PBMCs, or HLA-A2+TCL1− MCF-7 cells (Figure 6D-E). These results suggested that TCL1-specific CD8+ T cells that specifically recognize the autologous tumor cells could be generated from lymphoma patients.
TCL171-78 peptide-specific T cells can be generated from lymphoma patients. (A) PBMCs and tumor-infiltrating lymphocytes (TILs) derived from HLA-A2+ patients with CLL or FL were stimulated in vitro with TCL165-79. After 5 stimulations, IFN-γ production in response to TCL165-79 peptide was determined as described in Figure 3A. (B,E) Cytotoxic function of TCL165-79 peptide-specific T cells generated as described in panel A was tested against T2 cells pulsed with TCL165-79 peptide or HIV Gag77-85 peptide (B) or primary HLA-A2+ or HLA-A2− MCL or FL tumor cells (FL Tu) or tumor-free PBMCs from FL5 patient (FL5-PB) or HLA-A2− MCF-7 cell line (E). (C) Percentage of tetramer positive CD8+ T cells in each of the TCL1-specific T-cell lines generated from lymphoma patients is shown. (D) Expression of TCL1 in each of the primary lymphoma samples as determined by flow cytometry is shown. All cytotoxicity assays were performed in triplicate wells, and mean and standard deviation are shown. CLL1-PB-T indicates TCL1-specific CTL generated from peripheral blood T cells of CLL patient 1; FL5-PB-T, TCL1-specific CTL generated from peripheral blood T cells of FL patient 5; and FL5-TILs-T, TCL1-specific CTL generated from tumor infiltrating T cells of FL patient 5.
TCL171-78 peptide-specific T cells can be generated from lymphoma patients. (A) PBMCs and tumor-infiltrating lymphocytes (TILs) derived from HLA-A2+ patients with CLL or FL were stimulated in vitro with TCL165-79. After 5 stimulations, IFN-γ production in response to TCL165-79 peptide was determined as described in Figure 3A. (B,E) Cytotoxic function of TCL165-79 peptide-specific T cells generated as described in panel A was tested against T2 cells pulsed with TCL165-79 peptide or HIV Gag77-85 peptide (B) or primary HLA-A2+ or HLA-A2− MCL or FL tumor cells (FL Tu) or tumor-free PBMCs from FL5 patient (FL5-PB) or HLA-A2− MCF-7 cell line (E). (C) Percentage of tetramer positive CD8+ T cells in each of the TCL1-specific T-cell lines generated from lymphoma patients is shown. (D) Expression of TCL1 in each of the primary lymphoma samples as determined by flow cytometry is shown. All cytotoxicity assays were performed in triplicate wells, and mean and standard deviation are shown. CLL1-PB-T indicates TCL1-specific CTL generated from peripheral blood T cells of CLL patient 1; FL5-PB-T, TCL1-specific CTL generated from peripheral blood T cells of FL patient 5; and FL5-TILs-T, TCL1-specific CTL generated from tumor infiltrating T cells of FL patient 5.
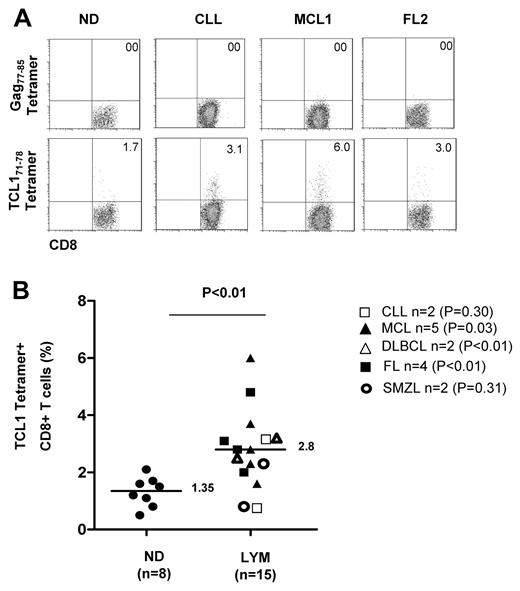
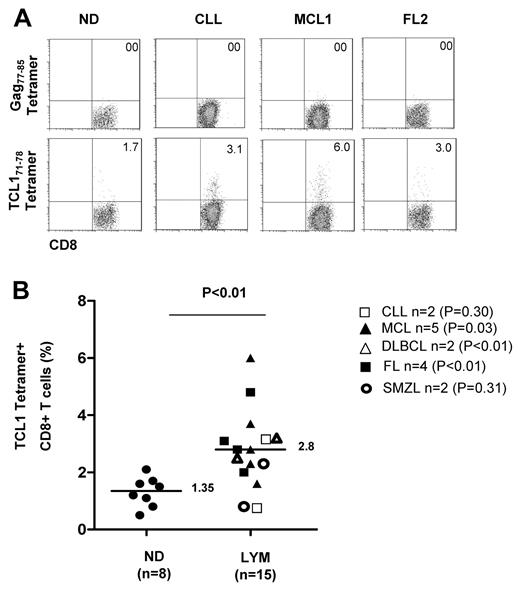
To determine whether TCL171-78 peptide-specific CD8+ T cells are naturally induced in lymphoma patients, we performed tetramer staining of PBMC derived from HLA-A2+ lymphoma patients at initial diagnosis (N = 15) and healthy donors (N = 8). Because the frequency of TCL1-specific CTL in the unstimulated PBMCs was below the threshold of detection by our tetramer assay, we performed 2 rounds of in vitro stimulation with TCL165-79 peptide. We found that the percentage of TCL171-78 tetramer-positive CD8+ T cells was significantly higher in patients (median, 2.8%; range, 0.88%-6%) compared with normal donor PBMC that were similarly stimulated in vitro (median, 1.35%; range, 0.38%-2.2%; (P < .01; Figure 7). When analyzed by each lymphoma subtype, TCL171-78 tetramer positive CD8+ T cells were significantly higher in MCL, DLBCL, and FL. These results suggested that TCL171-78 peptide-specific T cells are naturally induced in HLA-A2+ lymphoma patients. Moreover, these TCL171-78 tetramer-positive T cells had low levels of expression of coinhibitory molecules such as PD-1, CD160, CTLA-4, LAG-3, CD244, and BTLA (supplemental Figure 5). These results taken together with the data in Figure 6, suggest that the TCL1-specific T cells isolated from lymphoma patients were not exhausted.
TCL171-78 peptide-specific T cells are increased in lymphoma patients. PBMCs from HLA-A2+ ND (n = 8) or lymphoma patients (CLL, n = 2; MCL, n = 5; FL, n = 4; DLBCL, n = 2; and SMZL, n = 2) were stimulated in vitro twice with TCL165-79 peptide and stained with TCL171-78 tetramer or HIV Gag77-85 tetramer. Representative data of the tetramer staining (A) and results from all samples analyzed (B) are shown. P value on the top of graph represents comparison between normal donors and all lymphoma patient samples. P values to the right represent comparison between normal donors and patients from each lymphoma subtype. All P values were calculated by 2-tailed Student t test.
TCL171-78 peptide-specific T cells are increased in lymphoma patients. PBMCs from HLA-A2+ ND (n = 8) or lymphoma patients (CLL, n = 2; MCL, n = 5; FL, n = 4; DLBCL, n = 2; and SMZL, n = 2) were stimulated in vitro twice with TCL165-79 peptide and stained with TCL171-78 tetramer or HIV Gag77-85 tetramer. Representative data of the tetramer staining (A) and results from all samples analyzed (B) are shown. P value on the top of graph represents comparison between normal donors and all lymphoma patient samples. P values to the right represent comparison between normal donors and patients from each lymphoma subtype. All P values were calculated by 2-tailed Student t test.
Discussion
Our results indicate that TCL1 is a shared tumor-associated antigen for B-cell malignancies. Consistent with previous reports, we show that TCL1 is selectively expressed in normal B cells and their precursors in adults and is markedly hyperexpressed in most B-cell malignancies.13,15,16,30 The significant increase in protein expression for CLL, MCL, and FL (Figure 2B) despite a nonsignificant increase in mRNA (Figure 1C) may suggest posttranscriptional regulation of TCL1. More importantly, our results suggest that tumor cells naturally process and present TCL1 on their cell surface in the context of HLA-A2 molecules for recognition and killing by TCL1-specific CTL. TCL1-specific CTLs are not centrally deleted during thymic development because they can be generated from normal donors; however, high avidity CTLs may be deleted (supplemental Figure 3). Recent studies in prostate cancer37 and melanoma38 suggest that therapeutic vaccination with tissue-specific antigens that induce moderately avid T cells39 may induce clinical benefit. Furthermore, TCL171-78 peptide-specific CTL can be detected and expanded in vitro from the PBMCs and tumor-infiltrating lymphocytes of lymphoma patients, suggesting that TCL1 may be naturally immunogenic. However, the extent of the naturally induced TCL1-specific T cells may be insufficient to inhibit the growth of the tumor but could be augmented with immunotherapeutic strategies such as vaccines and adoptive T-cell therapy for enhanced antitumor effect and clinical benefit.
Although MHC class I molecules have been traditionally described to bind to peptides of 8 to 11 amino acids, recent reports suggest that peptides as long as 15 amino acids can bind to MHC class I molecules by bulging in the middle of the binding site or extending out of the MHC class I binding groove.31–35 Moreover, recent studies also demonstrated that long peptides may be more efficiently endocytosed, processed, and presented by antigen-presenting cells.40–43 By T2 binding assay, we observed that the TCL165-79 15 amino acid peptide bound to TAP-deficient T2 cells and was recognized by CTL when pulsed on to the T2 cells, indicating the direct binding of TCL165-79 (Figures 3C-E and 4A-C). Additional studies are needed to determine whether the antigen-presenting cells use cross-presentation pathway or cross-dressing pathway to process and present TCL165-79 peptide in vitro.44,45 Using a panel of truncated peptides, we identified TCL171-78 as the minimal epitope that bound to HLA-A2 and was recognized by TCL1-specific CTL. However, identification of the natural TCL1-derived HLA-A2 ligand would require isolation of MHC class I binding peptides from TCL1-expressing cells and analysis by mass spectrometry. Such studies may be necessary in the future for optimal design of the TCL1 vaccine formulation.
Our results, and those reported by others, suggest that TCL1 expression may be variable in tumor cells within each tumor.13,15,16,30 However, expression of low levels of TCL1 by the tumor cells may be sufficient because recognition by antigen-specific T cells requires only few MHC-peptide complexes on the cell surface.46 Furthermore, even if TCL1 is not present in all tumor cells within each tumor, lysis of TCL1-expressing tumor cells by TCL1-specific CTL may potentially induce epitope spreading and broaden the immune response against tumor cells with or without TCL1 expression. Indeed, a recent report demonstrates that epitope spreading develops in a high proportion of cancer patients who attain tumor regression after peptide-based cancer immunotherapy.47 Thus, the variable level of TCL1 expression in lymphoma cells, if present, may not be a limitation for using TCL1 as a shared lymphoma-associated antigen for therapeutic vaccine development.
The pattern of TCL1 expression seen in our studies is consistent with previous reports that suggested that TCL1 is expressed primarily in lymphoid tissues in precursor B and T cells and naive B cells in adults.15,16 Using real-time PCR, flow cytometry, and Western blotting, we found that TCL1 is hyperexpressed in tumor B cells compared with normal B cells in the peripheral blood. Consistent with this differential expression, we did not observe significant lysis of autologous B cells or PBMCs by TCL1-specific CTL. It is possible that the lysis of malignant B cells but not normal B-cell subsets despite that TCL1 expression also may be because of aberrant proteasome processing and MHC class I presentation of TCL1 peptides in malignant B cells compared with normal B cells, a phenomenon shown to occur with other tumor-associated antigens.48 Furthermore, our results suggest that hematopoietic stem cells do not express TCL1 and will not be targeted by TCL1-specific CTL. Nevertheless, it is possible that vaccination with TCL1 protein or TCL1-derived peptides may cause depletion of precursor, naive, and GC B cells. Although permanent depletion of B cells may result in hypogammaglobulinemia and increase the risk of infections, the morbidity can be minimized by prophylactic administration of intravenous γ-globulin as demonstrated in patients with X-linked agammaglobulinemia, a genetic disease characterized by lack of circulating B cells because of the mutation of Bruton tyrosine kinase.49 Depletion of normal B cells for several months after rituximab administration in patients with B-cell lymphoma also has not resulted in increased incidence of infections.50 Finally, TCL1-deficient mice had only modest deficiencies in B and T lymphopoiesis and slightly reduced IgG1 and IgG2b antibody production in response to a T-dependent antigen.51 Collectively, these reports suggest that targeting tumor B cells using TCL1-derived vaccine is likely to be safe and well tolerated.
Although idiotype vaccines have been found to be safe and immunogenic and induce clinical benefit in lymphoma patients, rapid progress in the development of idiotype vaccines has been hindered by the requirement for a tumor biopsy and the need to generate a custom-made product for each patient, a process that is expensive, laborious, and time-consuming. Identification of TCL1 as a shared lymphoma-associated antigen may lead to vaccine formulations that can be used in most lymphoma patients and are therefore easier and less costly to produce. Such shared lymphoma antigens would also facilitate rapid optimization of vaccination strategies by evaluating novel vaccine formulations, doses, and schedules, as well as novel adjuvants in clinical trials. Furthermore, identification of shared lymphoma-associated antigens would enable generation of autologous and allogeneic antigen-specific T cells for adoptive transfer.
In conclusion, our results suggest that TCL1 may serve as a shared tumor-associated antigen for therapeutic vaccine development in most of the common B-cell malignancies, including FL, CLL, MCL, DLCL, and SMZL. In this study, we identified TCL171-78 as the minimal HLA-A2–binding epitope but additional studies are needed to determine other HLA-class I and II binding peptides to broaden the clinical applicability of this tumor antigen to individuals that do not express HLA-A2. Alternatively, whole protein or genetic vaccines encoding TCL1 gene may be used without any restriction to HLA type. However, preclinical studies are necessary to design the optimal vaccine formulation and to determine the safety of TCL1 vaccination before evaluation in clinical trials.
There is an Inside Blood commentary on this article in this issue.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grant K23CA123149 (S.S.N.), the American Society of Clinical Oncology career development award (S.S.N.), the American Society of Hematology junior faculty scholar award in clinical/translational research (S.S.N.), and the Leukemia & Lymphoma Society Specialized Center research grant 7262-08 (L.W.K.). The tissue samples were provided by The University of Texas MD Anderson Cancer Center Lymphoma Tissue Bank. The Lymphoma Tissue Bank and the Flow Cytometry Core Facility are supported by The University of Texas MD Anderson Cancer Center support grant CA16672 (National Institutes of Health).
National Institutes of Health
Authorship
Contribution: J.W., H.J.P., and S.S.N. designed the study; J.W., S.R., F.C., H.J.P., R.S., and D.A.D. performed experiments; J.W., S.R., F.C., H.J.P., and S.S.N. analyzed the data; L.F., M.F., J.R., A.L., and L.W.K. provided patient samples; L.W.K. provided advice for the project; S.S.N. supervised all experiments; and J.W. and S.S.N. wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sattva S. Neelapu, Department of Lymphoma and Myeloma, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 0903, Houston, TX 77030; e-mail: sneelapu@mdanderson.org.