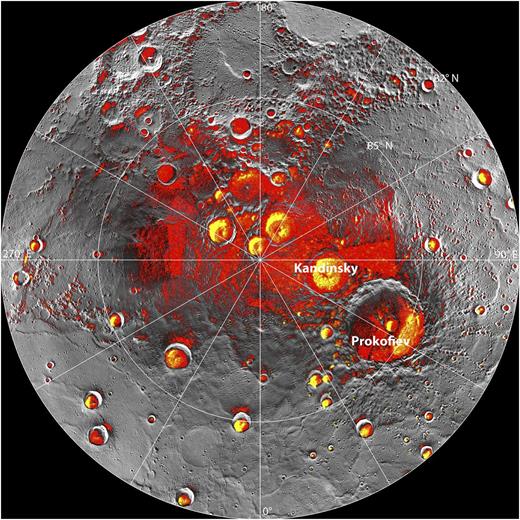
This image from NASA shows an overlay of Mercury’s new topographic map with the areas of Mercury’s pole that are in constant shadow (red) and those previously suggested to contain water-based ice from Earth-based reflected radio imaging from the Arecibo telescope (yellow). Together with other data from the MESSENGER spacecraft, these data support the view that Mercury harbors a significant amount of ice. Image credit: NASA/Johns Hopkins University Applied Physics Laboratory/Carnegie Institution of Washington/National Astronomy and Ionosphere Center, Arecibo Observatory.
This image from NASA shows an overlay of Mercury’s new topographic map with the areas of Mercury’s pole that are in constant shadow (red) and those previously suggested to contain water-based ice from Earth-based reflected radio imaging from the Arecibo telescope (yellow). Together with other data from the MESSENGER spacecraft, these data support the view that Mercury harbors a significant amount of ice. Image credit: NASA/Johns Hopkins University Applied Physics Laboratory/Carnegie Institution of Washington/National Astronomy and Ionosphere Center, Arecibo Observatory.
In early December of 2012, NASA reported that its MESSENGER spacecraft had imaged the entire surface of Mercury with pulses of infrared lasers resulting in the first detailed topographic map (see image). Mercury’s North Pole is in essentially constant darkness, and is therefore very cold, despite its proximity to our star. Earlier radar data from earth had suggested the hypothesis that cold northern craters might harbor water-based ice. Indeed, MESSENGER’s ability to shine a bright light from a short distance, collect thousands of images with the finest detectors, and relay volumes of data back to waiting scientists has transformed our view of our celestial sister. The data now overwhelmingly support the view that this otherwise scorching planet harbors ice.
In the same way, related technologies applied to our own field have intensely illuminated subtle changes in the genome that have had a disproportionate impact on the way cells behave, and misbehave. These new findings usher in a new era of malignant hematology.
In this issue of Blood, we launch a review series that will cover the exciting new findings linking epigenetic dysregulation to the pathogenesis of a number of hematologic diseases. The first review covers the fundamentals of epigenetic mechanisms and the role of some of the regulators in hematopoiesis. This will provide a framework for understanding their role in the clinical arena that will be covered in the following reviews:
The Role of Chromatin Modifiers in Normal and Malignant Hematopoiesis (Jill S. Butler and Sharon Y. R. Dent; The University of Texas MD Anderson Cancer Center Science Park)
Mechanisms of Epigenetic Deregulation in Lymphoid Neoplasms (Yanwen Jiang, Katerina Hatzi, and Rita Shaknovich; Weill Cornell Medical Center)
Mutations in Epigenetic Modifiers in the Pathogenesis and Therapy of Acute Myeloid Leukemia (Omar I. Abdel-Wahab and Ross L. Levine; Memorial Sloan-Kettering Cancer Center)
The Myelodysplastic Syndrome as a Prototypical Epigenetic Disease (Jean-Pierre Issa; Fels Institute, Temple University)
Epigenetic Mechanisms and Mixed Lineage Leukemia (Tobias Neff and Scott Armstrong; Memorial Sloan-Kettering Cancer Center)
Perspective and Future Directions for Epigenetics in Hematology (Margaret A. Goodell and Lucy A. Godley; Baylor College of Medicine and The University of Chicago)
Individual reviews will cover the specific associations of many of these regulators with lymphoid malignancies, myeloid malignancies, myelodysplastic syndrome (MDS), and mixed lineage leukemia (MLL)–associated disease. Our overall objective is to bring together the known, and highlight the unknown, to set forth some of the research and clinical goals for the next decade.
What is “epigenetics”?
“Epigenetics” refers to the heritable changes that impact cellular phenotype or physiology that do not occur at the level of alterations in the DNA sequence. Broadly, we can consider them semipermanent changes that effect a set of gene expression changes; however, they are reversible. A classic example is that of the ∼100 imprinted genes in the genome, of which only 1 allele is normally expressed, as determined by the gender of the parent from which each allele is derived. Imprinted genes are marked in a gender-specific manner during gamete formation by a still poorly understood mechanism that partly includes DNA methylation. After fertilization, the epigenetic marks on most genes are wiped clean, but for imprinted genes, the memory of the donating parent remains, which dictates the expression status of those genes for the lifetime of that generation—until the imprints are reset in the germline when the next generation of gametes are formed. This fascinating but rather abstruse mode of gene regulation is important in diseases such as Prader-Willi syndrome, in which uniparental disomy leads to an effectively null genic state due to “permanent” epigenetic repression of the remaining imprinted allele. The semipermanent nature of imprinting regulation illustrates the heritable but reversible nature of epigenetic marking.
In the case of regulation of imprinted genes, DNA methylation mediated by DNA methyltransferase 3a (DNMT3A) is critical in maintaining the marks established in the germline. In parallel, histones, which maintain the higher order structure of chromatin, can become modified by methylation and acetylation; the location and type of the particular modifications affects the manner in which the DNA is bound, and hence the accessibility to other gene-regulatory factors. These histone modifications are also considered semipermanent and are critical in maintaining the overall expression status of genes over many cell generations. The histone marks are deposited by proteins called “writers,” the marks are interpreted by “readers,” and they are capable of being “erased” when the expression status of a set of genes needs to be changed. Collectively, the proteins that execute these steps are called chromatin modifiers, and mutations in each class of these proteins can lead to various pathologic states, as described primarily for mice in this issue’s introductory review. In the subsequent reviews, we will see how their mutation can lead to hematologic malignancy.
A bright light on the field of epigenetics
Why this sudden surge of interest in epigenetics, when a decade ago malignant hematology was focused more on signaling pathways and tyrosine kinase inhibitors? The revolution comes from accumulating evidence that has developed through traditional means combined with the transformational effect that high-throughput DNA sequencing has had on virtually every aspect of biomedical research. Over the past ∼15 years or so, cloning of translocation breakpoints associated with malignancies have revealed the central role of some chromatin modifiers such as MLL proteins. Other accumulating evidence came from observations such as the effectiveness of 5-Aza-cytidine (5-Aza) for treatment of MDS patients; although we do not fully understand how 5-Aza works, it is well known to lead to DNA hypomethylation. Finally, DNA methylation has been known for decades to be abnormal in a variety of types of malignancies.
The tipping point however, that has brought all these disparate observations home, was the development of “next generation” sequencing. Massively high-throughput sequencing applied to hematologic malignancies has uncovered a myriad of new genes that are strongly implicated in the genesis of these diseases. Mutations in DNMT3A were initially linked to acute myeloid leukemia, but shortly thereafter to MDS, lymphoid malignancies, and several other disorders. Similarly, mutation in TET2, which modifies methylated cytosine to 5-hydroxymethylcytosine, is associated with a range of hematologic malignancies. Mutations in EZH2, which is critical for applying repressive histone marks, are now also associated with a variety of malignancies. The coming reviews in this series will focus on specific diseases and the collection of mutations in epigenetic regulators with which they are associated. Finally, we will try to build a broader perspective on what these findings, collectively, tell us from a scientific as well as clinical perspective.
Aside from identifying these proteins as central players in maintenance of normal hematopoiesis, where will this lead? Basic scientists will have years of work ahead to understand the mechanisms through which these specific sets of mutations lead to disease. More importantly for the patients, we have to think about how to interfere with the epigenetic state with new drugs. By definition, the epigenetic state is reversible—this should give us hope that new classes of drugs can be developed to revert the malignant epigenetic dysregulation.