In this issue of Blood, Baker and colleagues present the first study on genomic differences between multiple myeloma (MM) tumors derived from European Americans (EA) and African Americans (AA).1 They found a lower frequency of IgH translocations in AA compared with EA but otherwise similar genomic profiles.
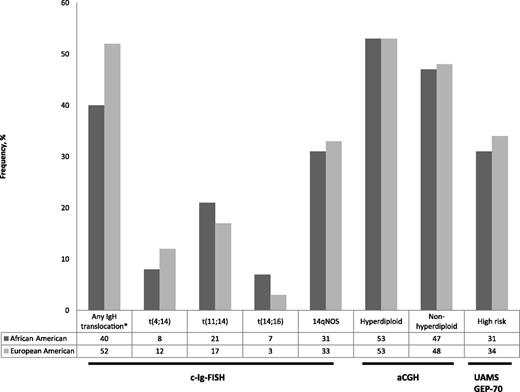
Frequency of genomic abnormalities in multiple myeloma tumors in African-Americans and European Americans (data from Baker et al).1 c-Ig-FISH, cytoplasmic light-chain fluorescence in situ hybridization; 14qNOS, no identifiable chromosome partner; aCGH, array comparative genomic hybridization; UAMS GEP-70, University of Arkansas Gene Expression Profiling 70 gene signature. *P = .032; all other comparisons not significant.
Frequency of genomic abnormalities in multiple myeloma tumors in African-Americans and European Americans (data from Baker et al).1 c-Ig-FISH, cytoplasmic light-chain fluorescence in situ hybridization; 14qNOS, no identifiable chromosome partner; aCGH, array comparative genomic hybridization; UAMS GEP-70, University of Arkansas Gene Expression Profiling 70 gene signature. *P = .032; all other comparisons not significant.
Myeloma has one of the most striking ethnic disparities in incidence and outcomes in cancer.2 Compared with incidence in EA, both MM and its precursor state, monoclonal gammopathy of undetermined significance (MGUS), are two- to threefold more common in AA. The clinical features of these plasma cell disorders are distinctly different between AA and EA. For example, AA patients with MGUS have lower monoclonal immunoglobulin (Ig) concentrations, a markedly lower frequency of IgM isotype, and a very high frequency (45% in AA compared with 33% in EA) of abnormal serum-free light-chain ratios.3 There is a strong suggestion that AA with MM may have more favorable outcomes than EA. Waxman and colleagues analyzed the National Cancer Institute SEER registries from 1973 to 2005 and demonstrated that the 5-year disease-specific survival for AA was 41.6% compared with 37.4% in EA (P < .001).4 However, in this analysis, AA did not experience the same degree of improvement in survival as EA diagnosed in more recent years. An analysis of the Center for International Bone Marrow Transplant Registry revealed that AA and EA treated uniformly with high-dose therapy and autologous stem cell transplantation have equivalent outcomes.5 The reasons for all of these differences are not clear, but taken together they suggest underlying biological differences between MM tumors in AA and EA patients.
Baker and colleagues sought to answer whether different genomic alterations between AA and EA with MM could account for these clinical observations. Molecular classification systems based on IgH translocations, chromosome content, and gene expression profiling have revealed genomic heterogeneity in MM that is associated with distinct clinical outcomes.6 Given the suggestion of better outcomes for AA compared with EA in clinical studies, a lower frequency of unfavorable genomic features might be expected in AA. They assembled a cohort of AA and EA MM tumors from the Eastern Cooperative Oncology Group trials E3A03 and E9487, the Multiple Myeloma Research Consortium (MMRC) Tissue Bank, and the Mayo Clinic. The available genomic data included IgH translocations by cIg–fluorescence in situ hybridization (FISH), array-based comparative genomic hybridization (aCGH), and gene expression profiling (GEP). The frequency of IgH translocations was 40% in AA compared with 52% in EA (P = .032). However, the frequency of the 3 most common partners—t(4;14), t(11;14), and t(14;16)—was not significantly different. There were similar frequencies of hyperdiploid and nonhyperdiploid MM tumors between EA and AA by aCGH. High-risk gene expression profiles using the University of Arkansas 70-gene expression (UAMS-70) signature were found in about one-third of both ethnicities. This study did not demonstrate a lower frequency of unfavorable genomic features in AA. A lower frequency of IgH translocations in AA is a new finding, but is it meaningful? Although the majority of IgH translocations [except t(11;14)] are associated with adverse outcomes, this study did not show significant differences in these translocations. At this point, a lower frequency of IgH translocations with an adverse prognosis as an explanation for improved outcomes in AA remains a hypothesis in need of further investigation.
The foremost limitation of this study is the very small proportion of MM tumors from AA, which hampers the ability to detect important differences. For example, in the E4A03 randomized phase III trial, nonwhite participants comprised only 60 of 445 patients (13%) and, of those, only 11 cases were available for cIg-FISH.7 Among tumors available for GEP from the MMRC, only 35 of 178 (16%) were from AA. Another limitation concerns the molecular classification itself. The molecular classification systems used are based on studies of predominantly EA patients. The UAMS-70 gene expression signature was developed from a population that was 90% EA. It is unknown whether the same spectrum of genomic findings is present in a representative population of MM tumors from AA. Another limitation is the heterogeneity of the assembled cohorts and the lack of clinical information. Finally, race and ethnicity influences cancer biology and clinical outcomes through many factors beyond the tumor genome, such as germline genetic variants, epigenomic changes, obesity, diet, inflammatory responses, the tumor microenvironment, and societal factors.8
Ethnic minorities are projected to comprise nearly half of the US population by 2050 (https://www.census.gov/population/projections). Recently, Ailawadhi reported results from SEER among EA, AA, Hispanics, and Asians with MM.9 They demonstrated that disease-specific survival was highest for Asians, followed in decreasing order by AA, EA, and Hispanics. Our understanding of disease biology in MM today is drawn from studies of predominantly EA and it is unknown whether these lessons will apply to other ethnic groups. The foremost challenge is the inclusion of ethnic minorities in clinical research. The MMRC has launched the CoMMpass study to perform whole-genome, whole-exome, and RNA sequencing of 1000 patients with newly diagnosed MM. This study is available at more than 50 academic and community sites across the United States and it is hoped will draw an ethnically diverse population of patients with MM to answer this important question. It is also critically important to study the myeloma precursor states (MGUS and smoldering multiple myeloma) and their progression to active MM in ethnically diverse populations.10 Such studies will inform our understanding of disease initiation and progression across ethnic groups.
Investigating biological differences among different racial and ethnic groups with MM will benefit all patients. Until results from adequately powered studies of ethnically diverse populations with MM are available, the findings of Baker and colleagues represent only a first step toward an understanding of multiethnic myeloma.
Conflict-of-interest disclosure: The author declares no competing financial interests.