Key Points
SOX4 is consistently expressed in ATL, is involved in ATL cell growth, and induces genes such as GCRK, NAP1, and HDAC8 in ATL.
FRA-2/JUND and SOX4 form an important oncogenic cascade in ATL, leading to upregulation of genes such as HDAC8.
Abstract
Previously, we have shown that an AP-1 family member, FRA-2, is constitutively expressed in adult T-cell leukemia/lymphoma (ATL) and, together with JUND, upregulates CCR4 and promotes ATL cell growth. Among the identified potential target genes of FRA-2/JUND was SOX4. Here, we examine the expression and function of SOX4 in ATL. SOX4 was indeed consistently expressed in primary ATL cells. FRA-2/JUND efficiently activated the SOX4 promoter via an AP-1 site. Knockdown of SOX4 expression by small interfering RNA (siRNA) strongly suppressed cell growth of ATL cell lines. Microarray analyses revealed that SOX4 knockdown reduced the expression of genes such as germinal center kinase related (GCKR), NAK-associated protein 1 (NAP1), and histone deacetylase 8 (HDAC8). We confirmed consistent expression of GCKR, NAP1, and HDAC8 in primary ATL cells. We also showed direct activation of the HDAC8 promoter by SOX4. Furthermore, siRNA knockdown of GCKR, NAP1, and HDAC8 each significantly suppressed cell growth of ATL cell lines. Taken together, we have revealed an important oncogenic cascade involving FRA-2/JUND and SOX4 in ATL, which leads to the expression of genes such as GCKR, NAP1, and HDAC8.
Introduction
Adult T-cell leukemia/lymphoma (ATL) is an aggressive mature CD4+ T-cell malignancy etiologically associated with human T-lymphotropic virus type 1 (HTLV-1).1 Previously, we and others have shown that most ATL cases strongly express CC chemokine receptor 4 (CCR4).2-4 We have further shown that FRA-2, a member of the AP-1 family transcription factors, is constitutively expressed in ATL and—together with another AP-1 family member, JUND—induces CCR4 expression and promotes cell growth in ATL.5 Furthermore, FRA-2/JUND induces the expression of various genes including proto-oncogenes such as c-MYB, MDM2, and BCL-6 in ATL.5 We have obtained similar results with cutaneous T-cell lymphomas (CTCLs), which also frequently express CCR4, albeit less strongly than ATL.6 These results suggest that FRA-2/JUND plays an important oncogenic role in mature T-cell malignancies expressing CCR4.
The list of potential target genes of FRA-2/JUND in ATL includes SOX4, a member of the Sry-related high mobility group box family of transcription factors.5 SOX4 is known to play important roles in various developmental processes including those that give rise to T cells and B cells.7,8 SOX4 is also frequently overexpressed in a variety of solid tumors and considered to be a potential oncogene.9-12 We therefore examined the expression and function of SOX4 in ATL. We demonstrated that SOX4 is indeed consistently expressed in ATL cell lines and primary ATL cells. We confirmed that FRA-2/JUND directly activates the SOX4 promoter via an AP-1 site. Furthermore, we demonstrated that SOX4 is involved in ATL cell growth and upregulates the expression of genes such as germinal center kinase related (GCKR),13 NAK-associated protein 1 (NAP1),14 and histone deacetylase 8 (HDAC8)15-17 in ATL. We confirmed direct activation of the HDAC8 promoter by SOX4. Furthermore, we demonstrated that GCKR, NAP1, and HDAC8 are each involved in ATL cell growth. Taken together, we have revealed an important oncogenic cascade involving FRA-2/JUND and SOX4 in ATL, which leads to the expression of downstream effector genes such as GCKR, NAP1, and HDAC8 in ATL.
Methods
Cells
All cell lines and primary cell samples used in this study were described previously.2,5,18 Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood samples obtained from healthy adult donors and acute ATL patients with a high leukemia cell count (>90%) using Ficoll-Paque (Amersham Biosciences Corp). Normal CD4+ T cells (purity, >96%) were further purified from PBMCs by negative selection using the IMagnet system (BD Pharmingen). Primary ATL cells and normal resting CD4+ T cells were stored in aliquots at –80°C and used for experiments without culture. Activated CD4+ T cells were prepared by culturing normal resting CD4+ T cells in the presence of Dynabeads Human T-Activator CD3/CD28 (Life Technologies) and 100 U/mL recombinant IL-2 (Shionogi) for 12 days. A written informed consent was obtained from each donor in accordance with the Declaration of Helsinki. This study was approved by the ethics committee of the Kinki University Faculty of Medicine.
RT-PCR
Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed as described previously5 using KOD FX DNA polymerase (TOYOBO, Osaka, Japan). In brief, the amplification conditions were denaturation at 94°C for 30 seconds (5 minutes for the first cycle), annealing at 60°C for 30 seconds and extension at 72°C for 30 seconds (5 minutes for the last cycle) for 30 cycles for FRA-2, JUNB, JUND, SOX4, GCKR, NAP1, and HDAC8, and 25 cycles for GAPDH. Amplification products were electrophoresed on 2% agarose gel and stained with ethidium bromide. The primers for FRA-2, JUND, JUNB, and GAPDH were described.5 The primers for SOX4, GCKR, NAP1, and HDAC8 are as follows: +5′-CACATCAAGCGACCCATGAAC-3′ (forward) and –5′-CCGGTACTTGTAGTCGGGGTAGT-3′ (reverse) for SOX4; +5′-TGAGGCAACGATGGAACAGTTAT-3′ (forward) and –5′-CATTTGTGGCAGCCTTTTGTATC-3′ (reverse) for GCKR; +5′-GGTTTGGAACAAGAGCTGGAACT-3′ (forward) and –5′-ACAGGCTTTCTTGATTGCAGTTG-3′ (reverse) for NAP1; +5′-GCACCATGGAGATGGTGTAGAAG-3′ (forward) and –5′-GACCACTGCTTTGGGATTAAAGG-3′ (reverse) for HDAC8.
Quantitative real-time PCR was performed as described previously.5 The primers and fluorogenic probes used for FRA-2, JUNB, JUND, GCKR, HDAC8, and GAPDH were obtained from the TaqMan kit (Applied Biosystems). We synthesized primers and fluorogenic probes for SOX4 and NAP1 as follows: +5′-CATGTCCCTGGGCAGCTT-3′ (forward), –5′-AGCCGGGCTCGAAGTTAAAA-3′ (reverse), and 5′-TCGTCGGCGCTCGACCGG-3′ (probe) for SOX4; +5′-TGAACTACTAAGAAAACTGAAAACCTCAA-3′ (forward), –5′-TCCAAGGTCTTCACTGCATCCT-3′ (reverse), and 5′-TGCAATCAAGAAAGCCTGTGCCCCT-3′ (probe) for NAP1.
Immunoblot analysis
Whole-cell lysates were prepared using radioimmunoprecipitation assay buffer (Thermo Fisher Scientific). Immunoblot analysis was performed as described previously.19 The following antibodies were used: rabbit anti-SOX4 (Abcam), monoclonal anti–α-tubulin (clone DM1A; Sigma-Aldrich), horseradish peroxidase–conjugated goat anti-rabbit immunoglobulin G (IgG) (Merck Chemicals), and goat anti-mouse IgG (GE Healthcare). Bands were visualized using the ECL plus detection kit (GE Healthcare).
Immunologic staining
These were performed as described previously.4 Briefly, cells were fixed, blocked, and reacted with rabbit anti-SOX4 (Abcam) or normal rabbit IgG (Beckman Coulter) and then with goat anti-rabbit IgG F(ab′)2–Alexa Fluor 546 (Invitrogen). After being counterstained with 4,6 diamidino-2-phenylindole (DAPI) (Invitrogen), cells were observed under a fluorescence microscope (BZ-8000; Keyence). Formalin-fixed paraffin-embedded biopsy tissues were obtained from archival tissue collections in the Department of Pathology, Kinki University Faculty of Medicine (ATL, n = 7; tonsillitis, n = 2). The tissue sections were heated 3 times for 5 minutes in target retrieval solution (DAKO) and treated with rabbit anti-SOX4 (Abcam) or normal rabbit IgG (DAKO) followed by biotin-labeled goat anti-rabbit IgG and the Vectastain ABC/HRP kit (Vector Laboratories). After the enzymatic development, sections were counterstained with Gill hematoxylin. Images were obtained using a Biozero BZ-8000 fluorescence microscope (objective, 40×) and BZ-II Analyzer software (Keyence).
Luciferase reporter assay
This assay was performed as described previously.5 Briefly, the major transcriptional start sites (+1) of SOX4 and HDAC8 were determined by the method of rapid amplification of the complementary DNA 5′ end (data not shown). The promoter regions of SOX4 (from –768 to +102 bp) and HDAC8 (from –712 to +126 bp) were amplified from human genomic DNA using PCR with the following primers: +5′-CATGCTAGCGACCCTTGCTTGGTTAAGAGAGT-3′ (forward) and –5′-CATCTCGAGCAGGAGTTCCTCCAGTGCAGACT-3′ (reverse) for SOX4; +5′-CATCTCGAGCAGATTTGCCAGAGTAAATTATTGAGA-3′ (forward) and –5′-CATAAGCTTACCAGCGACTGCCCACTGT-3′ (reverse) for HDAC8. The amplification products were digested with NheI and XhoI for SOX4 or XhoI and HindIII for HDAC8, and cloned into pGL3-basic luciferase reporter plasmid (Promega). Serial 5′-truncated promoter regions were similarly amplified using PCR with appropriate primers and cloned into pGL3. We also generated the SOX4 promoter fragment (from –768 to +102 bp) with a mutated AP-1 site (from CATGAGGAAGC to CATTTGGACTC) and the HDAC8 promoter fragment (from –712 to +126 bp) with a mutated SOX4 site (from AACAAGGA to ACCATGGA) using 2-step PCR with appropriate primers. The expression vectors for c-FOS, FRA-2, and JUND were described previously.5 The expression vectors for the spliced form of HTLV-1 bZIP factor (sHBZ) and SOX4 were generated as follows. We first constructed an expression vector, pIRES2/EGFP-EF1α, by inserting the EF1α promoter between the cytomegalovirus promoter and the multicloning sites of pIRES2/EGFP (Promega). sHBZ was amplified from an HTLV-1+ T-cell line TCL-Kan using PCR with the following primers: +5′-CATGAATTCGGCGTGGATGGCGGCCTCAGGGCTGTTTC-3′ (forward) and –5′-CATGGATCCTTATTGCAACCACATCGCCTCCAGC-3′ (reverse). Amplification products were digested with EcoRII and BamHI and cloned into pIRES2/EGFP-EF1α to make psHBZ-EGFP. Similarly, SOX4 complementary DNA was amplified from an ATL cell line ST1 using PCR with the following primers: +5′-CATGAATTCGAGGCCATGGTGCAGCAA-3′ (forward) and –5′-CATGGATCCTCAGTAGGTGAAAACCAG-3′ (reverse). Amplification products were digested with EcoRII and BamHI and cloned into pIRES2/EGFP-EF1α to make pSOX4-EGFP. The luciferase reporter gene driven by HTLV-1 long terminal repeat (LTR) (pGL3-HTLV-1-LTR) and the expression vector for HTLV-1 Tax (pHβPr.1-TAXMT2) were described previously.20 Using DMRIE-C transfection reagent (Invitrogen), cells (3 × 105) were transfected with 1.5 µg of a reporter plasmid, 0.5 µg of an expression plasmid for a transcription factor, and 0.3 µg of pSV-β-galactosidase (Promega). After 24 hours, assays were performed using a luciferase assay kit (Promega). Data were normalized with β-galactosidase activity, which served as an internal control for transfection efficiency.
ChIP assay
This was performed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) and following the manufacturer’s instructions. Briefly, cells (1 × 107) were cross-linked with 1% formaldehyde for 10 minutes at room temperature. Chromatins were digested to a size range between 150 and 900 bp using micrococcal nuclease. After brief sonication, cell lysates were obtained. The quality of chromatin samples was routinely checked by gel electrophoresis and ethidium bromide staining, which confirmed the amount and size range of chromatins. Chromatin samples were diluted 5-fold in chromatin immunoprecipitation (ChIP) buffer and incubated at 4°C overnight with 6 µg of anti-histone H3 (clone D2B12), a positive control provided in the kit (Cell Signaling Technology), anti–FRA-2 (sc-604), anti-JUND (sc-74), or normal rabbit IgG. Anti–FRA-2 and anti-JUND were obtained from Santa Cruz Biotechnology. Immune complexes were collected with protein G magnetic beads and incubated at 65°C for 2 hours to reverse the protein-DNA cross-links. DNAs were purified with a DNA-purification column. The AP-1 site of the SOX4 promoter was quantified using real-time PCR with the following primers and fluorogenic probe: +5′-GCACGCGGAGATTATTATTGC-3′ (forward), –5′-AACGCTTCCTCATGCCAAAC-3′ (reverse), and 5′-TCGGGTTCCAAGCCAATGGGAA-3′ (probe).
Cell proliferation assay
Small interfering RNAs (siRNAs) targeting FRA-2 (SI00420455), JUNB (SI03077445), JUND (SI00075985), and their control (1022064) were obtained from Qiagen. siRNAs targeting SOX4 (HSS106819), HDAC8 (HSS183330), GCKR (HSS117367), NAP1 (HSS127541), and their control (12935-145) were obtained from Invitrogen. Cells were transfected with siRNAs on Nucleofector (Lonza) using T solution and the O-17 program. The transfection efficiency of siRNAs was close to 95%, as determined using fluorescent siRNA. Cells overexpressing SOX4 were isolated 2 days after transfection with pSOX4-EGFP through sorting enhanced green fluorescent protein (EGFP)–expressing cells on FACSVantage (Becton Dickinson). In some experiments, cells were treated with broad-spectrum histone deacetylase (HDAC) inhibitors trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), which were purchased from Wako and Cayman Chemical, respectively. For cell proliferation assays, cells were seeded in a 96-well plate (0.5 × 104 cells per well) and viable cells were counted every 24 hours on FACSCalibur (Becton Dickinson) by gating out cells stained with propidium iodide.
Oligonucleotide microarray
Microarray analyses were performed using the Affymetrix GeneChip HG-U133 Plus 2.0 array (Affymetrix) as described previously.5 Briefly, ST1 cells were transfected with control siRNA or SOX4 siRNA as described in the previous section. After 48 hours, total RNAs were prepared and checked to be of good quality on Agilent 2100 Bioanalyzer (Agilent Technologies). All microarray data were submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo; accession no. GSE38021). The analysis was performed using BRB Array Tools software (version 3.3.0; http://linus.nci.nih.gov/BRB-ArrayTools.html) developed by Richard Simon and Amy Peng.
Statistical analysis
The Student t test was used to analyze differences between 2 groups. One-way analysis of variance with the Tukey post hoc test was used for multiple groups. All data were analyzed using GraphPad Prism (version 5.01; GraphPad Software). We considered P < .05 to be statistically significant.
Results
Expression of SOX4 in ATL
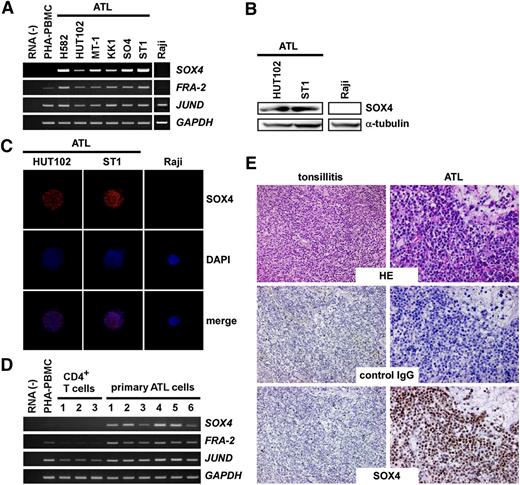
We first examined expression of SOX4 in ATL cell lines. As shown in Figure 1A, SOX4 was consistently expressed in ATL cell lines together with FRA-2 and JUND. Raji, a Burkitt lymphoma cell line, which was used as a negative control, expressed neither FRA-2 nor SOX4. We also confirmed SOX4 protein expression in 2 ATL cell lines (HUT102, ST1) by immunoblotting, which detected a 47-kDa band (Figure 1B), and by immunocytochemistry, which detected nuclear staining (Figure 1C). We next examined SOX4 expression in primary ATL samples. As shown in Figure 1D, PBMCs derived from acute type ATL patients (>90% leukemia cells) (n = 6) consistently expressed SOX4, FRA-2, and JUND at elevated levels (Figure 1D). In contrast, normal CD4+ T cells derived from healthy adult donors (n = 3) expressed SOX4 and FRA-2 only at low levels. Quantitative PCR confirmed elevated expression of SOX4 in ATL cell lines and primary ATL samples (supplemental Figure 1, available on the Blood website). Furthermore, immunohistochemical staining consistently revealed strong nuclear expression of SOX4 protein in infiltrating leukemic cells in skin lesions of ATL (n = 7) (Figure 1E). In contrast, anti-SOX4 hardly reacted specifically with normal tonsillar lymphocytes (n = 2) in comparison with control IgG (Figure 1E). These results confirm that SOX4 is consistently expressed at both messenger RNA (mRNA) and protein levels in ATL cells. Furthermore, strong nuclear staining of SOX4 is highly characteristic of ATL cells.
Expression of SOX4 in ATL. (A) RT-PCR analysis of ATL cell lines. Semiquantitative RT-PCR was performed to analyze expression of SOX4, FRA-2, and JUND in 6 ATL cell lines. Normal PBMCs treated with PHA for 3 days (PHA-PBMC) and Raji, a Burkitt lymphoma cell line, were also included. GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (B) Immunoblot analysis. Cell extracts prepared from 2 ATL cell lines and Raji were subjected to immunoblot analysis. After electrophoresis and electrotransfer, blotted filters were reacted with anti-SOX4 or anti–α-tubulin. The representative results from 3 separate experiments are shown. (C) Immunocytochemistry. Two ATL cell lines and Raji were used. Cells were fixed and stained with anti-SOX4 or control IgG. Cells were counterstained with DAPI. The representative results from 3 separate experiments are shown. Original magnification, ×800. (D) RT-PCR analysis of primary cells. Semiquantitative RT-PCR was performed to analyze expression of SOX4, FRA-2, and JUND in normal CD4+ T cells and primary ATL cells (>90% leukemia cells). GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (E) Immunohistochemistry. Tissue sections of inflamed tonsils (n = 2) and ATL skin lesions (n = 7) were stained with anti-SOX4 or control IgG. The representative results are shown. Original magnification, ×400. HE, hematoxylin-eosin; PHA, phytohemagglutinin.
Expression of SOX4 in ATL. (A) RT-PCR analysis of ATL cell lines. Semiquantitative RT-PCR was performed to analyze expression of SOX4, FRA-2, and JUND in 6 ATL cell lines. Normal PBMCs treated with PHA for 3 days (PHA-PBMC) and Raji, a Burkitt lymphoma cell line, were also included. GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (B) Immunoblot analysis. Cell extracts prepared from 2 ATL cell lines and Raji were subjected to immunoblot analysis. After electrophoresis and electrotransfer, blotted filters were reacted with anti-SOX4 or anti–α-tubulin. The representative results from 3 separate experiments are shown. (C) Immunocytochemistry. Two ATL cell lines and Raji were used. Cells were fixed and stained with anti-SOX4 or control IgG. Cells were counterstained with DAPI. The representative results from 3 separate experiments are shown. Original magnification, ×800. (D) RT-PCR analysis of primary cells. Semiquantitative RT-PCR was performed to analyze expression of SOX4, FRA-2, and JUND in normal CD4+ T cells and primary ATL cells (>90% leukemia cells). GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (E) Immunohistochemistry. Tissue sections of inflamed tonsils (n = 2) and ATL skin lesions (n = 7) were stained with anti-SOX4 or control IgG. The representative results are shown. Original magnification, ×400. HE, hematoxylin-eosin; PHA, phytohemagglutinin.
SOX4 is a direct target gene of FRA-2/JUND
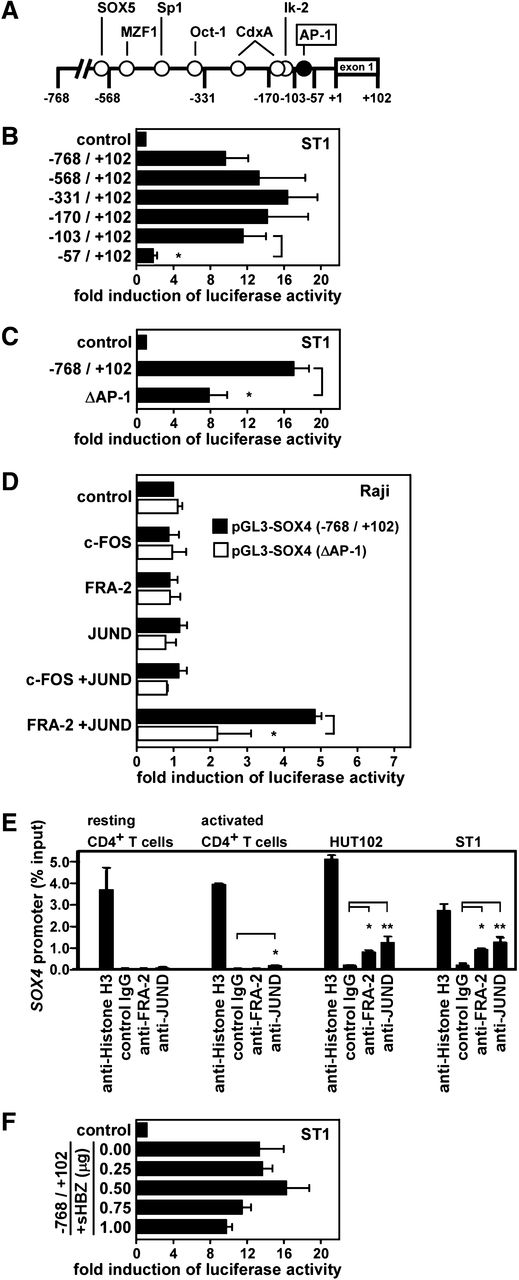
We next addressed whether SOX4 is a direct target gene of FRA-2 and JUND in ATL. Using siRNA knockdown and quantitative PCR, we confirmed that FRA-2 siRNA and JUND siRNA but not JUNB siRNA or control siRNA efficiently reduced SOX4 mRNA in ATL cell lines (supplemental Figure 2). We therefore examined the transcriptional control of the SOX4 promoter in ATL. Figure 2A shows the presence of potential cis elements in the SOX4 promoter region from –768 to –1 bp relative to the transcriptional start site. We inserted the DNA fragment from –768 to +102 bp and its successive 5′-truncated fragments into a luciferase reporter plasmid pGL3-basic and transfected these constructs to an ATL cell line ST1. As shown in Figure 2B, a strong promoter activity was observed until the –103-bp fragment, while the –57-bp fragment had little promoter activity. Thus, a major cis element(s) of the SOX4 promoter in ST1 cells was mapped within –103 to –57 bp. An AP-1 site was found in this region at –92 to –82 bp (Figure 2A). To directly address the role of this AP-1 site in the promoter activation, we generated a –768- to +102-bp construct with mutations in the AP-1 site (from CATGAGGAAGC to CATTTGGACTC). The SOX4 promoter activity in ST1 cells was indeed significantly (P < .05) reduced by the mutations in the AP-1 site (Figure 2C). The partial reduction could be due to incomplete inactivation of the AP-1 element or to the presence of other regulatory element(s) in this region. We next performed a reconstitution experiment using Raji, a Burkitt lymphoma cell line, which did not express SOX4 (Figure 1). Cells were cotransfected with the –768- to +102-bp construct of the SOX4 promoter with the wild-type or mutated AP-1 site and the expression vectors for c-FOS, FRA-2, and JUND, singly or in combination. As shown in Figure 2D, only the combination of FRA-2 and JUND activated the SOX4 promoter with the wild-type AP-1 site in Raji, while the combination of c-FOS and JUND had no such effect. Furthermore, the activation of the SOX4 promoter by FRA-2 and JUND was significantly (P < .05) reduced by the mutations in the AP-1 site. Again, the reduction was partial. Given that there were no other potential AP-1 sites in the promoter region used (Figure 2A), this might further support the notion that the inactivation of the AP-1 element was incomplete. Taken together, FRA-2/JUND indeed directly activates the SOX4 promoter at least partly via the AP-1 element at –92 to –82 bp.
Direct activation of the SOX4 promoter by FRA-2 and JUND. (A) A schematic depiction of the SOX4 promoter region from –768 to +102 bp with potential regulatory elements. (B) Deletion analysis. Luciferase reporter assays were performed using reporter constructs containing the SOX4 promoter fragment from –768 to +102 bp and its successive 5′-truncated fragments. The region from –103 to –57 bp was found to be necessary and sufficient for the reporter gene expression in ST1 cells. Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (C) Mutation analysis. Luciferase reporter assays were performed in ST1 cells using reporter constructs containing the SOX4 promoter fragment from –768 to +102 bp with an AP-1 site at –92 to –82 bp being wild-type or mutated (ΔAP-1, from CATGAGAAGC to CATTGGACTC). Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (D) Reconstitution experiment. Raji cells were cotransfected with pGL3-SOX4 (–768/+102) with the wild-type AP-1 site or mutated AP-1 site (ΔAP-1) and expression vectors for c-FOS, FRA-2, JUND, or control vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Promoter activation was shown as fold induction of luciferase activity in cells transfected with indicated AP-1 member expression vectors vs cells transfected with the control vector. Transfection efficiency was normalized by β-galactosidase activity. Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (E) ChIP assay. Chromatins prepared from normal resting and activated CD4+ T cells and 2 ATL cell lines (HUT102 and ST1) were fragmented and immunoprecipitated with anti-histone H3 (positive control), anti-FRA-2, anti-JUND, or control IgG (negative control). Real-time PCR was performed to quantify the AP-1 site of the SOX4 promoter in precipitated chromatin fragments relative to total input DNA (% input). Data are presented as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (F) Effect of HTLV-1 HBZ on the SOX4 promoter. ST1 cells were cotransfected with pGL3-SOX4 (–768/+102) and an expression vector for the spliced form of HBZ (sHBZ) or control vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Transfection efficiency was normalized by β-galactosidase activity. Data are presented as mean ± SEM of 3 separate experiments.
Direct activation of the SOX4 promoter by FRA-2 and JUND. (A) A schematic depiction of the SOX4 promoter region from –768 to +102 bp with potential regulatory elements. (B) Deletion analysis. Luciferase reporter assays were performed using reporter constructs containing the SOX4 promoter fragment from –768 to +102 bp and its successive 5′-truncated fragments. The region from –103 to –57 bp was found to be necessary and sufficient for the reporter gene expression in ST1 cells. Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (C) Mutation analysis. Luciferase reporter assays were performed in ST1 cells using reporter constructs containing the SOX4 promoter fragment from –768 to +102 bp with an AP-1 site at –92 to –82 bp being wild-type or mutated (ΔAP-1, from CATGAGAAGC to CATTGGACTC). Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (D) Reconstitution experiment. Raji cells were cotransfected with pGL3-SOX4 (–768/+102) with the wild-type AP-1 site or mutated AP-1 site (ΔAP-1) and expression vectors for c-FOS, FRA-2, JUND, or control vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Promoter activation was shown as fold induction of luciferase activity in cells transfected with indicated AP-1 member expression vectors vs cells transfected with the control vector. Transfection efficiency was normalized by β-galactosidase activity. Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (E) ChIP assay. Chromatins prepared from normal resting and activated CD4+ T cells and 2 ATL cell lines (HUT102 and ST1) were fragmented and immunoprecipitated with anti-histone H3 (positive control), anti-FRA-2, anti-JUND, or control IgG (negative control). Real-time PCR was performed to quantify the AP-1 site of the SOX4 promoter in precipitated chromatin fragments relative to total input DNA (% input). Data are presented as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (F) Effect of HTLV-1 HBZ on the SOX4 promoter. ST1 cells were cotransfected with pGL3-SOX4 (–768/+102) and an expression vector for the spliced form of HBZ (sHBZ) or control vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Transfection efficiency was normalized by β-galactosidase activity. Data are presented as mean ± SEM of 3 separate experiments.
To demonstrate the direct binding of FRA-2 and JUND to the AP-1 site of the SOX4 promoter in ATL cells, we next performed ChIP assays. Chromatins were prepared from 2 ATL cell lines (HUT102 and ST1) and normal resting and activated CD4+ T cells. The prepared chromatins were reacted with anti-histone H3 (positive control), anti-FRA-2, anti-JUND, or control IgG (negative control). Quantitative PCR was used to determine the amounts of DNA fragments containing the AP-1 site of the SOX4 promoter in immune complexes. As shown in Figure 2E, while anti-histone H3 precipitated the chromatins containing the AP-1 site of the SOX4 promoter from all samples with similar efficiencies, anti-FRA-2 and anti-JUND precipitated the chromatins containing the AP-1 site of the SOX4 promoter from 2 ATL cell lines but not from normal resting or activated CD4+ T cells. These results support the direct binding of FRA-2 and JUND to the AP-1 site of the SOX4 promoter in ATL.
HTLV-1 bZIP factor (HBZ), which is encoded by the minus strand of HTLV-1 provirus, is consistently expressed in primary ATL cells and considered to play an important role in ATL oncogenesis.21 Importantly, HBZ functions as a negative regulator of Tax-mediated HTLV-1 transcritpion.21 Recently, SOX4 was reported to be a possible downstream gene of HBZ.22 We therefore examined the effect of the splice form of HBZ (sHBZ), which is the major HBZ transcript in ATL,21 on the SOX4 promoter. In Raji cells, coexpression of sHBZ did not activate the SOX4 promoter at all but rather suppressed its activation by FRA-2 and JUND (data not shown). Similarly, cotransfection of the sHBZ expression vector did not activate the SOX4 promoter in ST1 cells but rather suppressed it at high plasmid doses (Figure 2F). In parallel, we confirmed that coexpression of sHBZ suppressed Tax-mediated activation of the HTLV-1 LTR promoter as reported previously (supplemental Figure 3).21 Thus, the sHBZ expression vector used was functional. Given that HBZ was reported to form a heterodimer with JUND,23 HBZ might compete with FRA-2 for JUND. Taken together, we concluded that SOX4 is not a direct target gene of HBZ.
Role of SOX4 in ATL cell growth
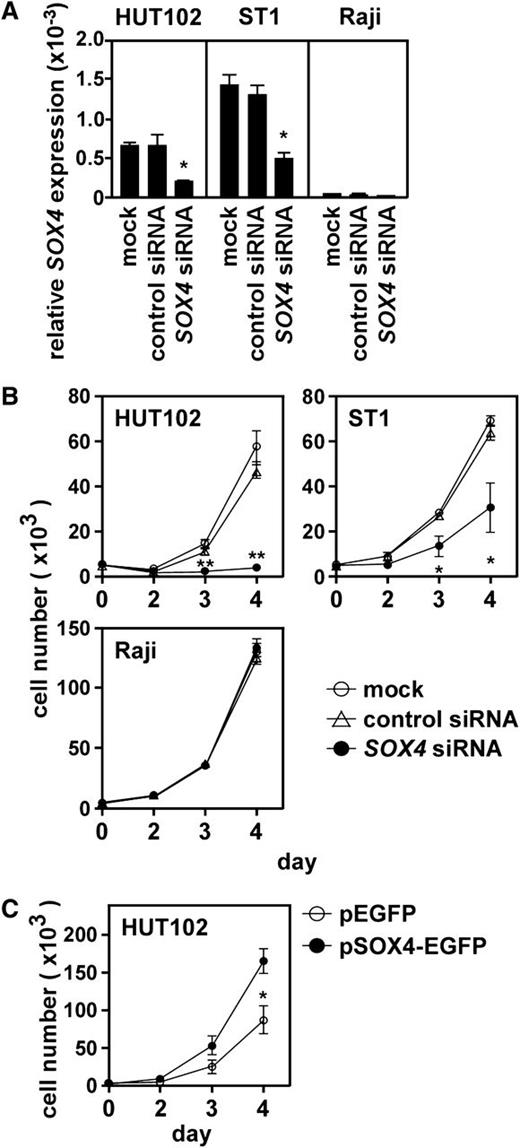
We next examined the role of SOX4 in ATL cell growth. Quantitative PCR confirmed that SOX4 siRNA but not control siRNA effectively reduced SOX4 mRNA in 2 ATL cell lines (HUT102 and ST1) (Figure 3A). Under these conditions, SOX4 siRNA but not control siRNA strongly suppressed cell growth of 2 ATL cell lines (Figure 3B). On the other hand, SOX4 siRNA had no effect on the cell growth of Raji (Figure 3B), which did not express SOX4 (Figure 1A). We next overexpressed SOX4 in HUT102, which only moderately expressed SOX4 (supplemental Figure 1). Overexpression of SOX4 strongly enhanced cell growth of HUT102 (Figure 3C). Taken together, these results clearly demonstrated that SOX4 is involved in ATL cell growth.
Role of SOX4 in ATL cell growth. (A) Effect of siRNA knockdown on SOX4 expression. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or SOX4 siRNA. After 48 hours, total RNA samples were prepared, and quantitative real-time PCR was performed. Data are shown as mean ± SEM of 3 separate experiments. *P < .05. (B) Effect of SOX4 siRNA on cell growth. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or SOX4 siRNA and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (C) Effect of SOX4 overexpression on cell growth. HUT102 cells were transfected with the control EGFP vector or pSOX4-EGFP. After 48 hours, cells expressing EGFP were sorted and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out dead cells stained with propidium iodide. Data are presented as mean ± SEM of 3 separate experiments. *P < .05.
Role of SOX4 in ATL cell growth. (A) Effect of siRNA knockdown on SOX4 expression. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or SOX4 siRNA. After 48 hours, total RNA samples were prepared, and quantitative real-time PCR was performed. Data are shown as mean ± SEM of 3 separate experiments. *P < .05. (B) Effect of SOX4 siRNA on cell growth. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or SOX4 siRNA and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (C) Effect of SOX4 overexpression on cell growth. HUT102 cells were transfected with the control EGFP vector or pSOX4-EGFP. After 48 hours, cells expressing EGFP were sorted and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out dead cells stained with propidium iodide. Data are presented as mean ± SEM of 3 separate experiments. *P < .05.
Identification of downstream target genes of SOX4 in ATL
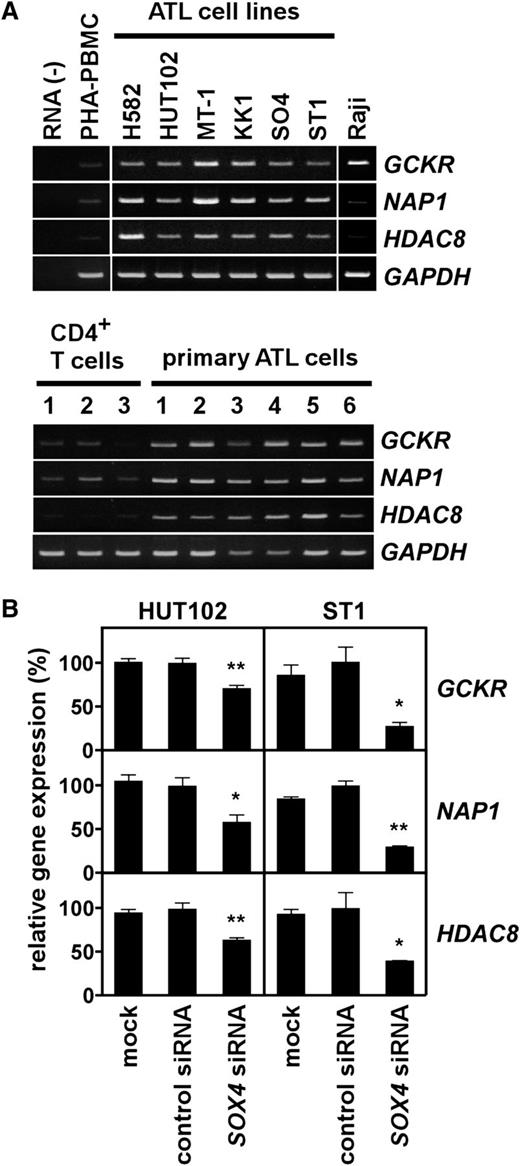
Given the strong effect of SOX4 on cell growth of ATL cell lines (Figure 3), we sought to identify downstream target genes of SOX4 in ATL. Using the Affymetrix high-density oligonucleotide microarray, we compared the gene expression profiles of ST1 cells transfected with control siRNA and SOX4 siRNA. Table 1 summarizes the top 50 genes whose expression was reduced >2-fold by SOX4 siRNA. To validate the microarray data, we selected 3 genes from Table 1 for further study; namely, GCKR (also called MAP4K5),13 NAP1 (also called AZ12),14 and HDAC8.15-17 These genes were chosen because they were likely to be important in ATL cell growth. As shown in Figure 4A, ATL cell lines and primary ATL samples consistently expressed GCKR, NAP1, and HDAC8. Quantitative PCR further confirmed elevated expression of HDAC8 in ATL cell lines and primary ATL samples (supplemental Figure 1). We also confirmed that SOX4 siRNA but not control siRNA significantly reduced the expression of GCKR, NAP1, and HDAC8 in 2 ATL cell lines (Figure 4B). These results in part validated our microarray data.
Expression of SOX4 target genes in ATL. (A) Gene expression analysis. Semiquantitative RT-PCR was performed to analyze expression of GCKR, NAP1, and HDAC8 in ATL cell lines and Raji (top panel), and in normal CD4+ T cells and PBMCs from ATL patients (>90% leukemia cells) (bottom panel). PHA-PBMC, normal PBMCs treated with PHA for 3 days. GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (B) Effect of SOX4 siRNA on target gene expression. Two ATL cell lines (HUT102 and ST1) were transfected with control siRNA or SOX4 siRNA. After 48 hours, total RNA samples were prepared. Quantitative real-time PCR was performed for GCKR, NAP1, HDAC8, and GAPDH. The expression levels were normalized by GAPDH. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01.
Expression of SOX4 target genes in ATL. (A) Gene expression analysis. Semiquantitative RT-PCR was performed to analyze expression of GCKR, NAP1, and HDAC8 in ATL cell lines and Raji (top panel), and in normal CD4+ T cells and PBMCs from ATL patients (>90% leukemia cells) (bottom panel). PHA-PBMC, normal PBMCs treated with PHA for 3 days. GAPDH was used as a loading control. The representative results from 3 separate experiments are shown. (B) Effect of SOX4 siRNA on target gene expression. Two ATL cell lines (HUT102 and ST1) were transfected with control siRNA or SOX4 siRNA. After 48 hours, total RNA samples were prepared. Quantitative real-time PCR was performed for GCKR, NAP1, HDAC8, and GAPDH. The expression levels were normalized by GAPDH. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01.
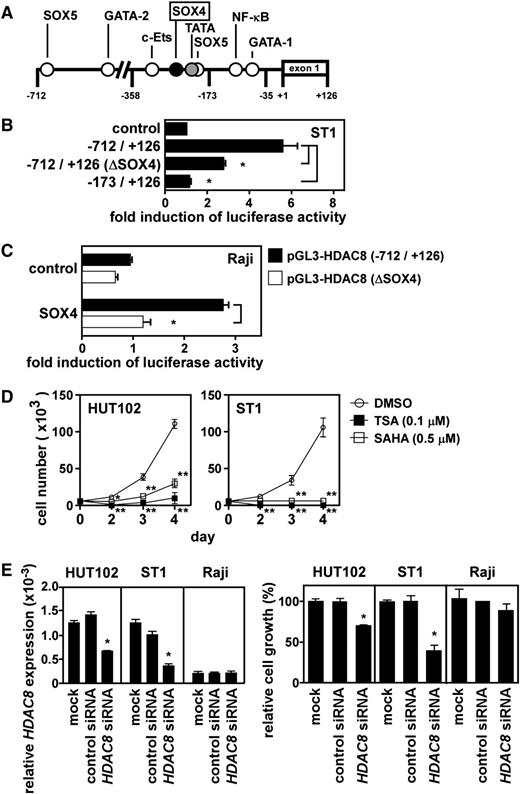
Direct activation of the HDAC8 promoter by SOX4
We further examined direct activation of the HDAC8 promoter by SOX4. Figure 5A depicts the promoter region of HDAC8 with potential cis elements. We inserted the DNA fragment from –712 to +126 bp relative to the transcriptional start site and its successive 5′-truncated fragments into pGL3-basic and transfected these constructs to ST1 cells for luciferase reporter assays. The results mapped a major regulatory element within –358 to –173 bp (data not shown). A potential SOX4 element was present at –243 to –236 bp. To directly address the role of this SOX4 element, we generated the –712- to +126-bp construct with mutations in the SOX4 element (from AACAAGGA to ACCATGGA). As shown in Figure 5B, the HDAC8 promoter activation in ST1 cells was significantly (P < .05) reduced by the mutations in the SOX4 element. Furthermore, cotransfection of the SOX4 expression vector in Raji cells efficiently activated the HDAC8 promoter with the wild-type SOX4 element but not that with the mutated SOX4 element (Figure 5C). Taken together, SOX4 indeed directly activates the HDAC8 promoter at least partly via the SOX4 element at –243 to –236 bp.
Direct activation of the HDAC8 promoter by SOX4. (A) A schematic depiction of the HDAC8 promoter region from –712 to +126 bp with potential cis regulatory elements. (B) Mutation analysis. Luciferase reporter assays were performed with the HDAC8 promoter fragment from –712 to +126 bp with the SOX4 site at –243 to –236 bp being wild-type or mutated (ΔSOX4, from AACAAGGA to ACCATGGA). Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (C) Reconstitution experiment. Raji cells were cotransfected with pGL3-HDAC8 (–712/+126) or pGL3-HDAC8 (–712/+126) ΔSOX4 and the SOX4 expression vector or control expression vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Promoter activation was expressed as fold induction of luciferase activity in cells transfected with the SOX4 expression vector vs cells transfected with the control expression vector. Transfection efficiency was normalized by β-galactosidase activity. Each bar represents mean ± SEM of 3 separate experiments. *P < .05. (D) Effect of broad-spectrum HDAC inhibitors on cell growth. ATL cell lines (HUT102 and ST1) were treated with TSA, SAHA, or vehicle (DMSO) and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (E) Effect of HDAC8 siRNA on cell growth. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or HDAC8 siRNA. After 48 hours, quantitative PCR was performed for HDAC8 and GAPDH. Levels of HDAC8 expression were normalized by GAPDH (left). Cells were also cultured in a 96-well plate at 0.5 × 104 cells per well. After 72 hours, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide (right). Data are presented as mean ± SEM of 3 separate experiments. *P < .05.
Direct activation of the HDAC8 promoter by SOX4. (A) A schematic depiction of the HDAC8 promoter region from –712 to +126 bp with potential cis regulatory elements. (B) Mutation analysis. Luciferase reporter assays were performed with the HDAC8 promoter fragment from –712 to +126 bp with the SOX4 site at –243 to –236 bp being wild-type or mutated (ΔSOX4, from AACAAGGA to ACCATGGA). Data are presented as mean ± SEM of 3 separate experiments. *P < .05. (C) Reconstitution experiment. Raji cells were cotransfected with pGL3-HDAC8 (–712/+126) or pGL3-HDAC8 (–712/+126) ΔSOX4 and the SOX4 expression vector or control expression vector as indicated. After 24 hours, luciferase assays were performed in triplicate. Promoter activation was expressed as fold induction of luciferase activity in cells transfected with the SOX4 expression vector vs cells transfected with the control expression vector. Transfection efficiency was normalized by β-galactosidase activity. Each bar represents mean ± SEM of 3 separate experiments. *P < .05. (D) Effect of broad-spectrum HDAC inhibitors on cell growth. ATL cell lines (HUT102 and ST1) were treated with TSA, SAHA, or vehicle (DMSO) and cultured in a 96-well plate at 0.5 × 104 cells per well. At indicated time points, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide. Data are shown as mean ± SEM of 3 separate experiments. *P < .05; **P < .01. (E) Effect of HDAC8 siRNA on cell growth. Two ATL cell lines (HUT102 and ST1) and Raji were transfected with control siRNA or HDAC8 siRNA. After 48 hours, quantitative PCR was performed for HDAC8 and GAPDH. Levels of HDAC8 expression were normalized by GAPDH (left). Cells were also cultured in a 96-well plate at 0.5 × 104 cells per well. After 72 hours, viable cells were counted on FACSCalibur by gating out cells stained with propidium iodide (right). Data are presented as mean ± SEM of 3 separate experiments. *P < .05.
Role of HDAC8, GCKR, and NAP1 in ATL cell growth
Recently, Hasegawa et al reported that a pan-HDAC inhibitor LBH589 effectively suppressed cell growth in ATL cell lines and primary ATL cells.24 Consistent with their report, broad-spectrum HDAC inhibitors TSA and SAHA efficiently suppressed cell growth in ATL cell lines (Figure 5D). In particular, ST1 cells were highly sensitive to the HDAC inhibitors. Furthermore, we demonstrated that HDAC8 siRNA but not control siRNA significantly suppressed cell growth of ATL cell lines (Figure 5E). HDAC8 siRNA had no such growth-suppressive effect on control Raji (Figure 5E). It was also notable that only partial reduction of HDAC8 expression efficiently suppressed ATL cell growth. In particular, ST1 cells were highly sensitive to the effect of HDAC8 knockdown. We further demonstrated that GCKR siRNA as well as NAP1 siRNA significantly suppressed cell growth of 2 ATL cell lines but not that of Raji (supplemental Figure 4).
Discussion
SOX4 is a member of the SOX (Sry-related high mobility group box) family of transcription factors that share homology in their DNA-binding domain, the high mobility group box.25 SOX4 is known to be involved in various biological processes including T- and B-cell development.7,8 Furthermore, overexpression of SOX4 was reported in a wide range of human cancers including breast, brain, lung, pancreatic, salivary gland, prostate, bladder, and ovarian cancers.9-12 The role of SOX4 in tumors may depend on the cellular context. For example, in the case of bladder cancer, strong expression of SOX4 significantly correlated with increased patient survival and overexpression of SOX4 induced apoptosis in cancer cells.9 Similarly, reduced expression of SOX4 was significantly correlated with metastasis and poor prognosis of melanoma patients.26 In sharp contrast, knockdown of SOX4 induced apoptosis in prostate and adenoid cystic cancer cells,10,27 and suppressed tumor growth and local metastasis in hepatocellular carcinoma.28 Recently, Pan et al reported a DNA damage sensor function of SOX4.29 They demonstrated that, upon DNA damage, SOX4 was quickly induced in parallel with p53 and promoted cell arrest and apoptosis in a p53-dependent manner.29 Thus, the strong upregulation of SOX4 in various solid tumors may in part reflect the increased DNA damages in such tumor cells. As for hematologic malignancies, Sox4 was identified as the gene most frequently associated with retroviral integration sites in a large panel of mostly B- and some T-cell lymphomas induced by retroviral insertion mutagenesis in AKXD and NFS.V+ mouse strains.30 This suggests that Sox4 is an important oncogene in B- and T-cell lymphomas. Sox4 was also demonstrated to promote mouse myeloid leukemia development in cooperation with Mef2c31 or in association with reduced expression of Sfpi1.32 Consistently, a significant negative correlation was observed in the expression levels of SOX4 and PU.1 (the human counterpart of Sfpi1) in human acute myeloid leukemia.32 Recently, a link between SOX4 and ATL was also suggested. A set of genes including SOX4 were shown to be up-regulated in the presence of HTLV-1 bZIP factor (HBZ),22 which is considered to play an important role in ATL oncogenesis.21 However, this study did not examine the mechanism of upregulation of SOX4 by HBZ or the role of SOX4 in ATL.
Previously, we have shown that the AP-1 family member FRA-2 is constitutively expressed in ATL and together with JUND up-regulates the expression of CCR4 and various other genes including several proto-oncogenes such as c-MYB, MDM2, and BCL6.5 SOX4 was also found among the potential downstream target genes of FRA-2/JUND in ATL.5 In the present study, we have shown that (1) SOX4 is indeed consistently expressed in primary blood-circulating as well as skin-infiltrating ATL cells (Figure 1), (2) FRA-2/JUND directly activates the SOX4 promoter via an AP-1 site (Figure 2), (3) HTLV-1 HBZ has no positive effect on the SOX4 promoter (Figure 2), and (4) SOX4 is involved in ATL cell growth (Figure 3). It is also notable that SOX4 is hardly expressed in normal CD4+ T cells in the blood or normal lymphoid tissues (Figure 1). Thus, SOX4 could be a novel diagnostic and therapeutic target molecule of ATL. Furthermore, by using an oligonucleotide microarray, we have identified potential downstream target genes of SOX4 in ATL (Table 1).
Among the genes in Table 1, we have chosen GCKR (also called MAP4K5),13 NAP1 (also called AZI2)14 and HDAC815-17 for further study. We focused on these genes because of their potential importance in ATL cell growth. We have confirmed their consistent expression in ATL cell lines and primary ATL samples (Figure 4). We have also shown direct activation of the HDAC8 promoter by SOX4 via a putative SOX4 site (Figure 5). We have further shown that HDAC8, GCKR, and NAP1 are all involved in cell growth of ATL cell lines (Figure 5, supplemental Figure 3). Although various genes were reported as potential target genes of SOX4 in bladder cancer,9 adenoid cystic carcinoma,27 and prostate cancer,10,33 GCKR, NAP1, and HDAC8 were not described in these previous studies and may thus be unique to ATL.
Mitogen-activated protein (MAP) kinases are activated in response to various cellular stimuli including cytokines and growth factors to mediate a wide range of cellular responses.34 There are 3 major groups of MAP kinases: the p38 MAP kinase family, the extracellular signal-regulated kinase family, and the c-Jun NH2-terminal kinase (JNK) family.34 The activation pathways of MAP kinases are composed of 3-tiered kinase cascades: MAPKs, MAPKKs (MAP2Ks), and MAPKKKs (MAP3Ks).34 MAP kinases are also known to be involved in the regulation of apoptosis and growth of hematologic malignancies.35 Germinal center kinase (GCK) related (GCKR, also called MAP4K5) is a member of the GCK family proteins. GCKR was originally reported as a major mediator of tumor necrosis factor–induced activation of stress-activated protein kinase (also called JNK).13 It has been shown that TRAF2 recruits and activates GCKR, which in turn activates stress-activated protein kinase/JNK.36 GCKR was also reported to be an important mediator of cellular transformation induced by Bcr-Abl, the oncogene associated in Philadelphia chromosome–positive myelogenous leukemia.37 Specifically, Bcr-Abl activates GCKR through Ras-dependent pathway to activate JNK.37 In acute myeloid leukemia, constitutive activation of JNK has been shown to correlate with treatment failure and increased multidrug resistance.38 Constitutive activation of JNK was also reported in ATL.39 Thus, the enhanced expression of GCKR may play a role in the constitutive activation of JNK in ATL, which in turn may activate c-JUN and JUND,40,41 leading to the constitutive activation of AP-1 in ATL.42
NAP1 (NAK-associated protein 1, also called AZI12) was initially identified as a protein interacting with NAK (IκB kinase–related kinase).14 NAP1 activates NAK to protect cells from tumor necrosis factor-α–induced apoptosis.14 NAP1 is also essential for double-stranded RNA–dependent production of interferon-β.43 The enhanced expression of NAP1 in ATL may thus play a role in the constitutive activation of nuclear factor–κB in ATL.44
HDACs are a group of enzymes whose primary functions are considered to be nucleosomal histone deacetylation, a major event that represses eukaryotic gene transcription. However, recent evidence has shown that HDACs are also involved in various cellular processes such as cell-cycle control, differentiation, and apoptosis by deacetylating a wide range of nonhistone proteins.45 HDACs are divided into 2 major groups: the zinc-dependent classical HDACs comprising class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9, and 10), and class IV (HDAC11) and the zinc-independent, NAD-dependent class III sirtuin enzymes.45 HDAC inhibitors have attracted much attention because of their potential application in cancer therapy.46,47 HDAC8 was originally cloned by 3 groups and shown to be expressed in various normal tissues and cancer cell lines with predominant nuclear localization in transfected cells.15-17 Subsequently, however, HDAC8 was specifically observed in cells with smooth muscle differentiation in normal human tissues, colocalized with the smooth muscle α-actin in the cytosol, and shown to be essential for smooth muscle cell contractile capacity.48,49 Importantly, an HDAC8-specific inhibitor, PCI-34051, was reported to induce caspase-dependent apoptosis through activation of phospholipase Cγ1 in T-cell lines derived from T-cell acute lymphoblastic leukemia or CTCLs but not in other hematopoietic or solid tumor cell lines.50 The reason for such selective activity is not clear but may be related to the dominant expression of phospholipase Cγ1 in T cells.50 Although PCI-34051 has not been tested for ATL, our present results strongly suggest that HDAC8-specific inhibitors such as PCI-34051 would be highly effective on ATL (Figure 5). In fact, a pan-HDAC inhibitor, LBH589, was already shown to have a potent antiproliferative effect on ATL.24 We also confirmed that the broad-spectrum HDAC inhibitors TSA and SAHA efficiently suppressed ATL cell growth (Figure 5). The enhanced expression of HDAC8 in ATL may in part account for the therapeutic effectiveness of HDAC inhibitors on ATL. The molecular function of HDAC8 in ATL remains to be seen.
In conclusion, we have demonstrated an important oncogenic cascade involving FRA-2/JUND and SOX4 in ATL, which induces target genes such as HDAC8, a promising molecular target of ATL and other T-cell malignancies.50 The aberrant expression of FRA-2 has also been frequently observed in CTCLs including anaplastic large-cell lymphoma.6,51 Indeed, we have found consistent expression of SOX4 and its downstream target genes such as HDAC8 in clinical samples of CTCLs (T.H., T.N., Naoki Oiso, Akira Kawada, and O.Y., manuscript in preparation). Thus, there may be a common oncogenic cascade involving FRA-2/JUND and SOX4 in mature T-cell malignancies such as ATL and CTCLs. The mechanism of aberrant expression of FRA-2 in ATL and CTCLs remains to be elucidated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Albert Zlotnik and Mitsugu Fujita for critical reading of the manuscript.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Authorship
Contribution: T.H., T.N., and O.Y. designed the research; T.H. and T.N. performed the research; T.A. and K.N. helped with the microarray analyses; and T.H. and O.Y. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.N. is Division of Chemotherapy, Kinki University Faculty of Pharmacy, Osaka, Japan.
Correspondence: Osamu Yoshie, Department of Microbiology, Kinki University Faculty of Medicine, 377-2 Ohno-Higashi, Osaka-Sayama, Osaka 589-8511, Japan; e-mail: o.yoshie@med.kindai.ac.jp.