In this issue of Blood, Liljeholm et al show that congenital dyserythropoietic anemia (CDA) type III is caused by a missense mutation in KIF23, which encodes a ubiquitous protein that regulates daughter cell separation during mitosis.1
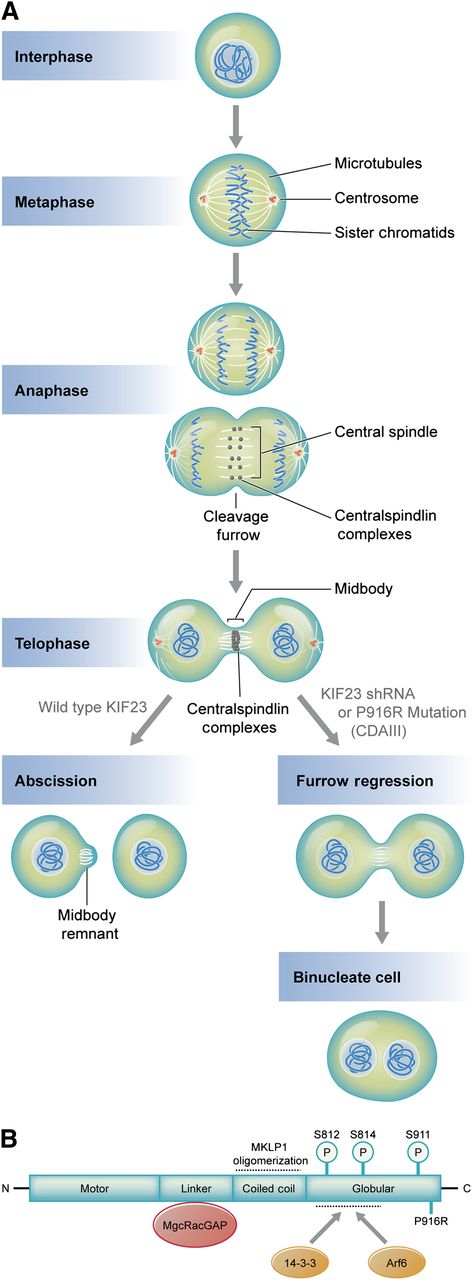
(A) Microtubule and DNA reorganization during mitosis and cytokinesis. Central spindle assembly during anaphase requires localization of centralspindlin a heterotetramer of MKLP1 and MgcRacGap/CYK-4, a GTPase activating protein. Compaction of the central spindle during telophase results in formation of the midbody, a process requiring centralspindlin. Normally, the midbody is cleaved in a process termed abscission, yielding 2 mononuclear cells. MKLP1 deficiency or aberrant MKLP1, as occurs in CDA III, results in cleavage furrow regression and ultimately binucleate cells. Subsequent rounds of failed cytokinesis could result in erythroid cells with up to 12 nuclei, as observed in CDA III. (B) Domain interactions of MKLP1 (adapted from White and Glotzer7 ). The linker region and coiled coil domains mediate MgcRacGAP interaction and promote oligomerization, respectively. Oligomerization promotes activity by enhancing interactions with microtubules. Interaction with 14-3-3 proteins inhibits cytokinesis by recruiting centralspindlin away from microtubules at the central spindle or midbody. This interaction is positively and negatively regulated by phosphorylation. Phosphorylation of S812 enhances activity by inhibiting interaction with 14-3-3, whereas S814 phosphorylation promotes 14-3-3 binding. Phosphorylation of S911 enhances MKLP1 activity by preventing its premature import into the nucleus during cytokinesis. Interaction with ARF6 GTPase enhances activity by competing with 14-3-3 binding. The P916R CDA III mutation described by Liljeholm et al, which impairs MLKP1 activity, is indicated at the C-terminus. Professional illustration by Kenneth X. Probst.
(A) Microtubule and DNA reorganization during mitosis and cytokinesis. Central spindle assembly during anaphase requires localization of centralspindlin a heterotetramer of MKLP1 and MgcRacGap/CYK-4, a GTPase activating protein. Compaction of the central spindle during telophase results in formation of the midbody, a process requiring centralspindlin. Normally, the midbody is cleaved in a process termed abscission, yielding 2 mononuclear cells. MKLP1 deficiency or aberrant MKLP1, as occurs in CDA III, results in cleavage furrow regression and ultimately binucleate cells. Subsequent rounds of failed cytokinesis could result in erythroid cells with up to 12 nuclei, as observed in CDA III. (B) Domain interactions of MKLP1 (adapted from White and Glotzer7 ). The linker region and coiled coil domains mediate MgcRacGAP interaction and promote oligomerization, respectively. Oligomerization promotes activity by enhancing interactions with microtubules. Interaction with 14-3-3 proteins inhibits cytokinesis by recruiting centralspindlin away from microtubules at the central spindle or midbody. This interaction is positively and negatively regulated by phosphorylation. Phosphorylation of S812 enhances activity by inhibiting interaction with 14-3-3, whereas S814 phosphorylation promotes 14-3-3 binding. Phosphorylation of S911 enhances MKLP1 activity by preventing its premature import into the nucleus during cytokinesis. Interaction with ARF6 GTPase enhances activity by competing with 14-3-3 binding. The P916R CDA III mutation described by Liljeholm et al, which impairs MLKP1 activity, is indicated at the C-terminus. Professional illustration by Kenneth X. Probst.
CDAs have puzzled hematologists for years. Bone marrow red blood cell precursors from affected patients are dysmorphic with multiple nuclei and fail to develop effectively into mature red blood cells.2 Although several CDA genes have been identified, how their altered function leads to erythroblast multinuclearity is obscure.
In this issue of Blood, Liljeholm et al show that CDA III, a rare autosomal dominant disorder, is caused by a missense mutation in KIF23, which encodes mitotic kinesin-like protein 1 (MKLP1), a key component of the apparatus that ensures faithful separation of cells during late mitosis.1 This study identifies, for the first time, a potential mechanism by which a CDA mutation causes multinuclearity, a characteristic finding in this group of disorders.
The CDAs are broadly classified into 3 types (I, II, and III) based on distinctive morphological and clinical features, although rare “atypical” forms exist.2 Positional cloning identified mutations in the CDAN1 and SEC23B genes as causative for CDAs I and II, respectively.2 CDANI encodes codanin 1, a poorly understood protein that may facilitate histone assembly into chromatin and regulate the cell cycle. SEC23B encodes a protein involved in endoplasmic reticulum vesicle trafficking. Other forms of CDA are caused by mutations in GATA1 and KLF1 genes, which encode essential erythroid transcription factors. CDA III was the first CDA to be described (in 1951), although its genetic etiology has escaped detection until just now.3 In the 1990s, Lind and colleagues used linkage studies to map the CDA III gene to chromosome 15q21-25 in a large multigenerational Swedish pedigree.4 Liljeholm et al narrowed the region to 2.5 Mb by using current human genome data to reanalyze haplotypes defined by the Lind study. Through next-generation sequencing of the minimal region, Liljeholm et al identified a c.2747C>G (p.P916R) missense mutation in the KIF23 gene. This mutation co-segregates with CDA III in the original Swedish pedigree and an American one as well.
MKLP1 is a well-studied kinesin-like motor protein that facilitates completion of mitosis.5,6 During anaphase, the region between sister chromatids reorganizes to form the central spindle, consisting of arrayed microtubules and a host of associated proteins, including centralspindlin, a heterotetramer of MKLP1 and MgcRacGAP/CYK-4, a GTPase activating protein (see figure).7 A cleavage furrow is established via formation of a contractile ring. In telophase, the central spindle narrows into a thin intercellular bridge, or midbody, which is eventually severed to resolve 2 daughter cells via a process termed abscission. During these events, centralspindlin organizes microtubules, regulates the activity of small GTPases involved in forming the contractile ring, connects the central spindle/midbody to the plasma membrane, and recruits various proteins that promote abscission.6,8 Disruption of any of these processes can cause regression of the cleavage furrow, resulting in a binuclear, tetraploid cell (see A in figure). Multiple rounds of failed cytokinesis could cause more than 2 nuclei to accumulate in a single cell, as is observed in CDA III. Indeed, more than 60 years ago, Wolff and von Holfe speculated that multinucleate cells of CDA III result from completed nuclear division with failed cytokinesis.3 Liljeholm et al used time-lapse microscopy to show that this is precisely what occurs in HeLa cells when MKLP1 P916R replaces the normal protein.
The current study is interesting and satisfying because it provides a direct mechanistic link between the extensively studied biology of mitosis and the multinuclearity of CDA. More generally, this work illustrates how modern genetics can rapidly and efficiently identify human disease causing mutations. Of course, many questions remain. For example, if MKLP1 is an essential component for mitosis in all cells, why does the P916R mutation produce a phenotype that is largely (though not completely) erythroid-restricted? This situation is similar to Diamond Blackfan anemia in which haploinsufficiency of ribosomal proteins affects tissue development selectively. These clinical observations indicate that redundant biochemical pathways exist for fundamental cellular processes and that different tissues exhibit unique thresholds for pathology when these processes are perturbed. It is also possible that tissue-specific forms of MKLP1 exert unique functions, perhaps 1 of which is to facilitate the specialized cell divisions that occur during erythroid maturation. Indeed, MKLP1 is expressed at particularly high levels in red blood cell precursors where KIF23 is bound by the essential erythroid transcription factors GATA1 and TAL1 (http://biogps.org/#goto=genereport&id=9493; http://genome.ucsc.edu). Additionally, Liljeholm et al identified a novel MKLP1 alternative splice isoform that is enriched in hematopoietic tissues including peripheral blood, bone marrow, and spleen. However, the consequences of MKLP1 P916R may not be entirely erythroid-specific, because CDA III patients exhibit increased risk for multiple myeloma, a potential oncogenic consequence of failed cytokinesis.9,10
It is also of interest to better understand how the P916R mutation impairs MKLP1 function. Is this simply haploinsufficiency for MKLP1 or does this particular mutation have a specific dominant negative effect? This mutation occurs at the carboxyl terminus of the protein, a region that participates in several important protein interactions, some of which are regulated by phosphorylation (see B in figure). Future studies should ascertain how the P916R mutation affects these phosphorylation events, the assembly of the centralspindlin complex, and/or its recruitment of partner proteins to the central spindle and midbody.
Finally, this study may provide a toehold for understanding how other disorders cause erythroid multinuclearity. For, example, it is possible that the CDA-associated transcription factors GATA1 or KLF1 regulate MKLP1 expression during erythroid maturation. Likewise, SEC23B and/or codanin 1 may function in pathways that regulate MKLP1 or associated proteins. Myelodysplastic syndromes and erythroleukemias also exhibit erythroblast multinuclearity, perhaps by altering processes that occur at the central spindle and midbody. It is possible that polyploidy associated with these acquired disorders contributes to their malignant transformation. In these ways, the study by Liljeholm et al raises interesting possibilities as to how a variety of diverse inherited and acquired anemias may converge on a common pathway to cause erythroid multinuclearity.
Conflict-of-interest disclosure: The authors declare no competing financial interests.