To the editor:
A recent genome-wide association study of childhood acute lymphoblastic leukemia (ALL) by Xu et al1 has identified a novel susceptibility locus encompassing BMI1-PIP4K2A at chr10p12.31-12.2, bringing the number of known ALL risk loci to 5 (including ARID5B, IKZF1, CEBPE, and CDKN2A).2-4 We sought to confirm and refine the BMI1-PIP4K2A association using single nucleotide polymorphism (SNP) array data from 297 Hispanic children with B-cell ALL (B-ALL) and 454 Hispanic control children from the California Childhood Leukemia Study.5
Of 6 genome-wide significant BMI1-PIP4K2A SNPs reported by Xu et al,1 their most strongly associated variant (rs7088318) was not significantly associated with B-ALL in our array data (P = .21). Intriguingly, the rs7088318 association approached significance when analyses were limited to hyperdiploid B-ALL (P = .08), the most common cytogenetic subgroup. We subsequently imputed genotypes using 1000 Genomes data (supplemental Methods) and saw a similar association pattern for their 5 remaining SNPs (supplemental Table 2), suggesting that this region may specifically confer risk for the hyperdiploid B-ALL subtype.
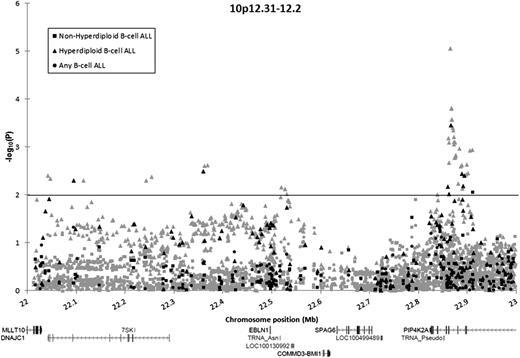
In total, 131 directly genotyped and 1371 imputed SNPs in the BMI1-PIP4K2A region were analyzed. Association tests were stratified into 3 groups: B-ALL, hyperdiploid B-ALL, and nonhyperdiploid B-ALL. No SNP was significantly associated with the risk for B-ALL in unstratified analyses (P > .01). A total of 8 array SNPs and 43 imputed SNPs were associated with hyperdiploid B-ALL risk (P < .01) (Figure 1; supplemental Table 3). The array SNP most significantly associated with hyperdiploid B-ALL risk was rs10764338 (odds ratio [OR], 2.39; 95% confidence interval [CI], 1.48-3.87; P = 3.5 × 10−4), but this SNP was not significantly associated with an increased risk for B-ALL in unstratified analyses (OR, 1.34; 95% CI, 0.97-1.85; P = .078).
Association of SNPs in the BMI1-PIP4K2A locus with B-ALL among Hispanic children, by ploidy. Association of 131 directly genotyped SNPs (black) and 1371 imputed SNPs (gray) with the risk for B-ALL, adjusted for sex, age, and the first 5 principal components. Circles denote associations for children with B-ALL compared with control children. Squares denote associations for children with nonhyperdiploid B-ALL compared with control children. Triangles denote associations for children with hyperdiploid B-ALL compared with control children.
Association of SNPs in the BMI1-PIP4K2A locus with B-ALL among Hispanic children, by ploidy. Association of 131 directly genotyped SNPs (black) and 1371 imputed SNPs (gray) with the risk for B-ALL, adjusted for sex, age, and the first 5 principal components. Circles denote associations for children with B-ALL compared with control children. Squares denote associations for children with nonhyperdiploid B-ALL compared with control children. Triangles denote associations for children with hyperdiploid B-ALL compared with control children.
One SNP (rs1750761) was associated with nonhyperdiploid ALL at P < .01 but had effects in opposite directions in analysis of nonhyperdiploid cases (OR, 0.55) vs analysis of hyperdiploid cases (OR, 1.62). In case-by-case comparisons, on comparison of hyperdiploid cases (N = 97) with nonhyperdiploid cases (N = 157), our top directly genotyped SNP from case-control analyses (rs10764338) was again significantly associated with an increased risk for hyperdiploid B-ALL (OR, 2.47; 95% CI, 1.42-4.29; P = 9.8 × 10−4).
Although risk loci in ARID5B have been shown to confer a greater risk for hyperdiploid B-ALL vs other subtypes,2,4 the risk loci in BMI1-PIP4K2A reported here are the first to be exclusively associated with this ALL subtype. Our results confirm that variation at chr10p12.31-12.2 confers a risk for childhood ALL and further indicates that these variants distinguish hyperdiploid B-ALL from other subtypes. Fine-mapping via SNP imputation refined the association peak to a ∼35kb region in PIP4K2A, which includes a rare variant (rs142846483; minor allele frequency in controls = 0.011) conferring an exceptionally high risk for hyperdiploid B-ALL among Hispanic children (OR, 15.15; 95% CI, 4.57-50.21 P = 8.8 × 10−6).
The risk variants identified by Xu et al do not affect protein coding, although they found that the rs7088318 risk allele was associated with increased PIP4K2A messenger RNA expression.1 Our most strongly associated SNPs were also intronic, suggesting that variants in PIP4K2A that confer risk for hyperdiploid B-ALL may be regulatory in nature, as has been observed in fine-mapping studies of other neoplastic diseases.6,7
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors gratefully acknowledge the clinical investigators at the following collaborating hospitals for help in recruiting patients: University of California Davis Medical Center (Dr Jonathan Ducore), University of California San Francisco (Drs Mignon Loh and Katherine Matthay), Children’s Hospital of Central California (Dr Vonda Crouse), Lucile Packard Children’s Hospital (Dr Gary Dahl), Children’s Hospital Oakland (Dr James Feusner), Kaiser Permanente Roseville (formerly Sacramento) (Drs Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs Carolyn Russo, Alan Wong, and Denah Taggar), Kaiser Permanente San Francisco (Dr Kenneth Leung), and Kaiser Permanente Oakland (Drs Daniel Kronish and Stacy Month).
Contribution: K.M.W., A.J.d.S., and J.L.W. conceived and designed the study; L.F.B., P.A.B., A.P.C., G.V.D., L.H., C.M., K.M.W., and J.L.W. assisted in assembling the data; K.M.W. analyzed and interpreted the data; A.J.d.S. and K.M.W. wrote the manuscript; and all authors critically reviewed and edited the manuscript for intellectual content and gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kyle M. Walsh, University of California, San Francisco Helen Diller Family Cancer Center, Box 0520, 1450 3rd St HD276, San Francisco, CA; e-mail: kyle.walsh@ucsf.edu.
References
Author notes
K.M.W. and A.J.d.S. contributed equally to this work.