Key Points
Defective Ras oncoproteins initiate T-ALL.
Murine T-ALLs lacking PTEN have gene expression profiles similar to human early T-cell precursor ALL and are resistant to MEK inhibition.
Abstract
Reversing the aberrant biochemical output of oncogenic Ras proteins is one of the great challenges in cancer therapeutics; however, it is uncertain which Ras effectors are required for tumor initiation and maintenance. To address this question, we expressed oncogenic K-RasD12 proteins with “second site” amino acid substitutions that impair PI3 kinase/Akt or Raf/MEK/ERK activation in bone marrow cells and transplanted them into recipient mice. In spite of attenuated signaling properties, defective K-Ras oncoproteins initiated aggressive clonal T-lineage acute lymphoblastic leukemia (T-ALL). Murine T-ALLs expressing second site mutant proteins restored full oncogenic Ras activity through diverse mechanisms, which included acquiring novel somatic third site KrasD12 mutations and silencing PTEN. T-ALL cell lines lacking PTEN had elevated levels of phosphorylated Akt, a gene expression pattern similar to human early T-cell precursor ALL, and were resistant to the potent and selective MEK inhibitor PD0325901. Our data, which demonstrate strong selective pressure to overcome the defective activation of PI3 kinase/Akt and Raf/MEK/ERK, implicate both Ras effector pathways as drivers of aberrant growth in T-ALL and further suggest that leukemia cells will deploy multiple mechanisms to develop resistance to targeted inhibitors in vivo.
Introduction
Somatic RAS mutations encode oncogenic proteins that accumulate in an active signaling conformation.1-3 Although the biophysical properties of Ras oncoproteins render them exceedingly challenging targets for rational drug discovery, recent data suggest that this might be feasible.4 There is also intensive interest in inhibiting Ras-regulated kinase cascades in cancer, particularly the Raf/MEK/ERK and PI3K/Akt/mTOR pathways.1,5 To maximize the efficacy of either therapeutic strategy, it is essential to identify Ras effectors required for cancer initiation and maintenance.
Expressing Ras oncoproteins with “second site” amino acid substitutions that mediate binding to individual effectors is a robust approach for investigating this question, complementing the use of small-molecule inhibitors while avoiding potential confounding problems such as off-target activities and unpredictable levels of kinase inhibition in vivo.6-8 Previous studies in fibroblasts and epithelial cells support the idea that simultaneous activation of PI3K, Raf, and Ral-GDS is essential for Ras-induced tumorigenesis.1-3,6-8 Determining requirements for hyperactive signaling through different effector pathways in hematologic cancers has translational implications, given the high prevalence of somatic RAS mutations.4,9
A glycine-to-aspartic acid substitution at codon 12 (D12) is the most common KRAS mutation found in human cancer. Here we show that oncogenic K-RasD12 proteins containing second site substitutions at glutamate 37 (K-RasD12/G37) or tyrosine 64 (K-RasD12/G64) are impaired for activating Raf/MEK/ERK and PI3K signaling, respectively. Expressing either mutant protein in mouse bone marrow cells unexpectedly deregulated the growth of myeloid progenitors in vitro and initiated aggressive T-lineage acute lymphoblastic leukemia (T-ALL) in vivo. These leukemias displayed biochemical properties that correlated with responses to targeted inhibitors and with distinct secondary genetic alterations, including acquired third site mutations within KrasD12 transgenes. We conclude that aberrant PI3K/Akt and Raf/MEK/ERK signaling contribute to T-ALL growth and suggest that leukemia cells will deploy both on-target and off-target mechanisms to overcome targeted inhibitors.
Methods
Kras expression constructs
Wild-type (WT) Kras mouse cDNA was cloned into the pENTR/D-TOPO vector (Invitrogen). We used a QuikChange site-directed mutagenesis kit (Stratagene) to introduce point mutations and Gateway technology (Invitrogen) to clone Kras cDNAs into the pDEST12.2 vector (Invitrogen) and into a murine stem cell virus (MSCV) vector containing a green fluorescent protein (GFP) cassette driven by an internal ribosome entry site (IRES) downstream of the Kras sequence (MIG [MSCV-IRES-GFP]). For some in vitro experiments, we used MSCV vectors in which GFP was fused to the NH2 end of Kras (MSCV-GFP-Kras).10
Retroviral transduction and progenitor colony assays
The University of California, San Francisco, Committee on Animal Research approved procedures involving mice. E14.5 C57Bl/6 fetal livers were isolated as described.1,5,11 MIG plasmids were cotransfected with packaging plasmids into 293T cells using Lipofectamine2000 (Invitrogen) and viral 3-day transfer, inoculum of 3×105 cells (3T3) fibroblasts and fetal liver cells transduced with supernatant. GFP-positive (GFP+) fetal liver cells were isolated on a FACSAria (BD Biosciences) and seeded in methylcellulose (M3231; StemCell Technologies) containing recombinant mouse granulocyte macrophage colony-stimulating factor (GM-CSF). Colony-forming-unit granulocyte macrophage (CFU-GM) colonies were counted by indirect microscopy after 8 days. All cytokines were from Peprotech unless otherwise noted.
Studies in COS-7 and S49 cells
COS-7 cells were transfected with pDEST12.2 plasmids. After 24 hours, medium was changed to Iscove modified Dulbecco medium (IMDM; University of California, San Francisco, Core; starve) or IMDM + 20% fetal bovine serum (FBS; Hyclone; basal). Cells were harvested 24 hours later (starve) or after exposure to 50 ng/mL of epidermal growth factor for 5 minutes (stimulated). Unstarved cells were harvested in parallel (basal). S49 cells were transduced with MIG plasmids and sorted as described earlier. GFP+ cells were starved for 2 hours in Dulbecco’s modified Eagle medium H21 (University of California, San Francisco, Core) before harvest. Immunoblotting was performed as previously described.6-8,12 All immunoblot antibodies were from Cell Signaling except total Akt (Biosource).
Transduction/transplantation procedure
WT Balb/c mice were injected with 150 mg/kg 5-fluorouracil 4 days before euthanasia. Bone marrow cells were collected into IMDM + 20% FBS; cultured in StemSpan SFEM (Stem Cell) with 15% FBS, 100 ng/mL interleukin 11 (IL-11; R&D Technologies), 100 ng/mL stem cell factor, 50 ng/mL fms-related tyrosine kinase 3, 50 ng/mL IL-6, and 10 ng/mL IL-3; transduced with retroviral supernatant after 24 to 72 hours; and transplanted 24 hours later. Male WT Balb/c mice were lethally irradiated with a single 850-cGy dose and retroorbitally injected with transduced cells 2 to 3 hours later. Secondary recipients of established leukemias received a single 500-cGy dose. Blood cells were counted by Hemavet (Drew Scientific), and smears were stained with Wright Giemsa (Sigma-Aldrich). The University of California, San Francisco, Mouse Pathology Core analyzed organs. For fluorescence-activated cell sorting (FACS) analysis, cells were resuspended after red cell lysis in Hanks balanced salt solution + 3% FBS and Fc block and then stained with antibodies against myeloid/erythroid (PE-Cy7-Mac1, PacBlue-Gr1, PE-CD71, and APC-Ter119), T-cell (PE-Cy7-CD3, PE-CD8, and APC-CD4), B-cell (PE-Cy7-B220, PacBlue-CD19) ,and stem cell (PE-Sca1, APC-ckit) markers (BD Biosciences). Data were acquired with LSRII (BD Biosciences), using FACSDiva software, and analyzed with FlowJo (Tree Star).
DNA purification and Southern blotting
Hematopoietic tissues were lysed with 100 mM Tris-HCl at pH 8.5, 5 mM EDTA pH at 8.0, 200 mM NaCl, and 0.2% sodium dodecyl sulfate. Genomic DNA was digested with EcoRI and then hybridized with a sequence-verified GFP probe, as previously described.12
T-ALL cell lines
Single-cell suspensions from bone marrow, thymus, or spleen of sick mice were used to general T-ALL cell lines, as previously described.12 After serial passage, cells were harvested from basal culture conditions or after 24 hours of starvation in Dulbecco’s modified Eagle medium H21 and lysed (50 mM Tris-HCL at pH 8.0, 150 mM NaCl, 5 mM MgCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate). Ras was immunoprecipitated with H-Ras (sc-259) and probed with pan-RasAsp-12 (both Santa Cruz Biotechnology) to detect mutant Ras. Ras-GTP was immunoprecipitated with Raf1-RBD agarose conjugate beads (Millipore). Total Ras (Millipore) was measured before immunoprecipitation.
Biochemical analysis of fetal liver cells
E14.5 fetal liver cells transduced as described earlier were resuspended in Hanks balanced salt solution + 3% FBS and Fc block and then stained with Pac Blue-Mac1. Sorted GFP+, Mac1+ cells were immunoblotted with anti-Ras. For phospho-flow analysis, unsorted cells were resuspended in starve (IMDM + 1% BSA) or basal (IMDM + 20% FBS) media and then incubated for 2 hours at 37°C. Fixed and permeabilized cells were incubated with Fc Block and then stained with Pac Blue-Mac1 and Alexa 647-pAkt (Thr308), anti-pERK plus PE secondary (Jackson Immune Research), or anti-pS6 plus PE. FACS data were collected as done earlier.
Proliferation and apoptosis assays
T-ALL cell lines were plated at a density of 30 000/100 µL in 96-well plates. The drug was added in varying concentrations in triplicate. After 48 hours, 20 µL CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) was added, and the plates were incubated for 4 hours. Plates were read according to the manufacturer's instructions. Growth curves were established as percentages of maximal growth in dimethylsulfoxide, and half maximal inhibitory concentration (IC50) values were calculated. For apoptosis assays, 50 000 cells were plated in 1 µM PD0325901. After 48 hours, the cells were collected, fixed with paraformaldehyde, and stained with an antibody against cleaved Caspase-3 (BD Biosciences). FACS data were acquired as described earlier.
Expression profiling
Gene expression data were generated with GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) with signals normalized to the trimmed average of 500 in the MAS 5.0 algorithm. Probe sets with absent calls for all samples were excluded, and probe set signals were variance-stabilized by adding 32 and log2 transformation. Statistical analyses were performed using R 2.11.0 (http://r-project.org), Bioconductor 2.6,13 and Spotfire Decision Site 9.1.1 (Tibco). Supervised analysis to detect differentially expressed genes between PI3K activated and nonactivated groups was performed using limma14 with estimation of false-discovery rate.15 Genes with a false-discovery rate below 20% were considered significantly differentially expressed and used to assess pathway enrichment in Database for Annotation, Visualization and Integrated Discovery version 6.7.16,17 Gene expression profiling of 52 T-ALL samples was performed with Affymetrix GeneChip HT HG-U133+ PM arrays, with signals normalized by robust multi-array average algorithm. The correlation between mouse PI3K activation and early T-cell precursor (ETP) ALL expression was examined with gene set enrichment analysis18 on human ETP against non-ETP, using the repository of gene sets available at MSigDB version 3.0 and the top 100 up-regulated mouse genes in PI3K activation.
Statistical analysis
Statistics were analyzed with Prism 4 (GraphPad). Kaplan-Meier survival curves were compared by log-rank test with a 2-tailed P value. Percentage maximum growth = (mean colonies in 3 replicate plates)/(mean colonies in plates with no GM-CSF) × 100. Two-tailed t tests were used to compare all other data sets.
Results
KrasD12 substitutions at codons 37 and 64 retain GM-CSF hypersensitivity
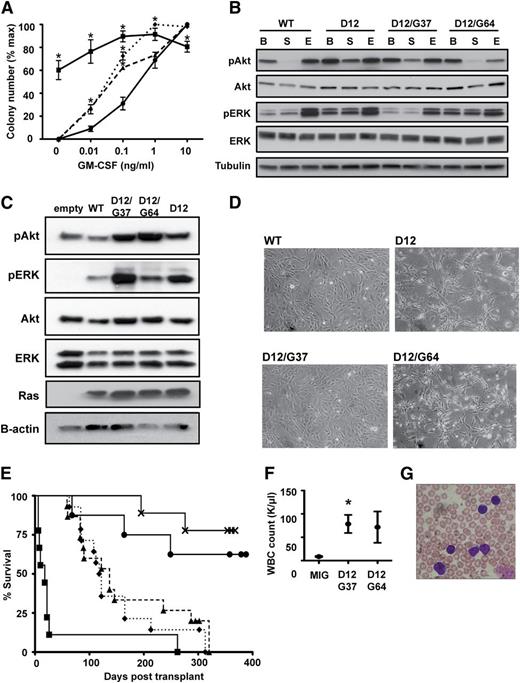
We constructed MSCV-IRES-GFP retroviruses encoding WT K-Ras, K-RasD12, or K-RasD12 proteins containing amino acid substitutions within the switch I and switch II domains (supplemental Table 1, available on the Blood Web site).2,6,7 After infection, GFP+ mouse fetal liver cells were plated in methylcellulose. As expected, cells infected with the MSCV-KrasD12-IRES-GFP vector formed CFU-GM colonies without added cytokines, and those that grew in plates containing GM-CSF were abnormally large and monocytic.1,11 Second site mutant K-Ras proteins that interact with a single class of effectors6,7 did not perturb colony growth (supplemental Table 1 and supplemental Figure 1A). However, KrasD12 alleles encoding glycine substitutions at either glutamate 37 (K-RasD12/G37) or tyrosine 64 (K-RasD12/G64) induced modest hypersensitivity, characterized by an increase in the number of CFU-GM colonies formed at low concentrations of GM-CSF (Figure 1A). These data indicate that K-RasD12/G37 and K-RasD12/G64 exhibit gain of function compared with WT K-Ras but are biologically less activated than oncogenic K-RasD12.
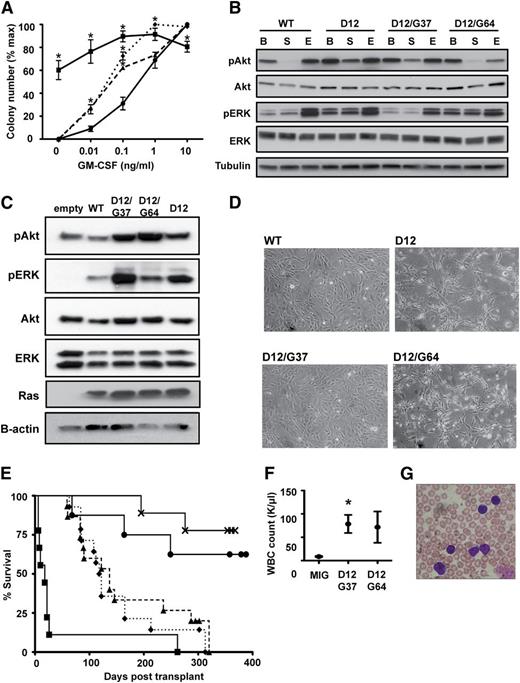
Second site KrasD12 mutant alleles induce hypersensitive myeloid progenitor growth and initiate T-ALL. (A) CFU-GM formation by fetal liver cells expressing WT Kras (●), KrasD12 (▪), KrasD12/G37 (▲), or KrasD12/G64 (♦). Mean ± standard error of the mean (SEM) of 5 independent experiments is shown. Data points marked with an asterisk (*) are significantly different from WT by paired, 1-tailed t test (P < .05). (B) Signaling in transfected COS-7 cells under basal (B), starved (S), and EGF-stimulated (E) conditions. (C) Signaling in transduced S49 cells under starved conditions. These cells were infected with MSCV-GFP-Kras vectors and sorted to equalize Ras expression levels. GFP-K-Ras fusion proteins run at a different molecular weight than endogenous Ras proteins. (D) Morphology of 3T3 cells expressing WT Kras, KrasD12, KrasD12/G37, or KrasD12/G64. Note that KrasD12 and KrasD12/G64 induce morphologic changes. (E) Survival of lethally irradiated WT mice transplanted with bone marrow cells transduced with MIG vector (×; n = 8) or MIG vectors expressing WT Kras (●; n = 10), KrasD12 (▪; n = 9), KrasD12/G37 (▲; n = 15), or KrasD12/G64 (♦; n = 14). (F) White blood cell (WBC) counts at death in recipients of bone marrow transduced with MIG vector (n = 8), KrasD12/G37 (n = 15), or KrasD12/G64 (n = 14) viruses. Data plotted as mean ± SEM, with an asterisk (*) indicating data points significantly different from WT by unpaired, 1-tailed t test (P < .05). (G) Peripheral blood smear showing blast morphology in a mouse with T-ALL.
Second site KrasD12 mutant alleles induce hypersensitive myeloid progenitor growth and initiate T-ALL. (A) CFU-GM formation by fetal liver cells expressing WT Kras (●), KrasD12 (▪), KrasD12/G37 (▲), or KrasD12/G64 (♦). Mean ± standard error of the mean (SEM) of 5 independent experiments is shown. Data points marked with an asterisk (*) are significantly different from WT by paired, 1-tailed t test (P < .05). (B) Signaling in transfected COS-7 cells under basal (B), starved (S), and EGF-stimulated (E) conditions. (C) Signaling in transduced S49 cells under starved conditions. These cells were infected with MSCV-GFP-Kras vectors and sorted to equalize Ras expression levels. GFP-K-Ras fusion proteins run at a different molecular weight than endogenous Ras proteins. (D) Morphology of 3T3 cells expressing WT Kras, KrasD12, KrasD12/G37, or KrasD12/G64. Note that KrasD12 and KrasD12/G64 induce morphologic changes. (E) Survival of lethally irradiated WT mice transplanted with bone marrow cells transduced with MIG vector (×; n = 8) or MIG vectors expressing WT Kras (●; n = 10), KrasD12 (▪; n = 9), KrasD12/G37 (▲; n = 15), or KrasD12/G64 (♦; n = 14). (F) White blood cell (WBC) counts at death in recipients of bone marrow transduced with MIG vector (n = 8), KrasD12/G37 (n = 15), or KrasD12/G64 (n = 14) viruses. Data plotted as mean ± SEM, with an asterisk (*) indicating data points significantly different from WT by unpaired, 1-tailed t test (P < .05). (G) Peripheral blood smear showing blast morphology in a mouse with T-ALL.
Glu-37 and Tyr-64 substitutions selectively impair Ras effector activation
The crystal structures of Ras bound to Raf, Ral-GDS, and PI3K suggest how amino acid substitutions at codons 37 and 64 of Ras selectively impair effector interactions.2,19 Glu-37 is expected to form 2 hydrogen bonds with Raf, and substitutions at Glu-37 impair Raf binding while only weakly affecting Ral binding.20 Glu-37 also interacts with a basic residue on the p110α subunit of PI3K that is not present on p110δ or p110γ. Tissue-specific differences in PI3K isoform expression likely underlie the variable effects of H-RasD12/G37 expression on PI3K signaling.7,8 Notably, blood cells express high levels of p110δ and p110γ, suggesting that Ras proteins containing substitutions at Glu-37 will retain the ability to activate PI3K signaling in hematopoietic tissues.21 Substitutions at switch II residue Tyr-64 selectively disrupt the interaction between Ras and PI3K, as Raf and Ral-GDS do not bind to this effector domain.22 Consistent with these data, mutating amino acids within p110α that contact Tyr-64 abolished Ras binding.23 Supplemental Table 1 summarizes what has been learned about substitutions at codons 37 and 64 from in vitro binding assays, structural studies, and biochemical investigation. Notably, Ras proteins containing E37G and Y64G substitutions retain the ability to bind to and activate Ral-GDS.6,22
We expressed K-RasD12/G37 and K-RasD12/G64 in COS-7 cells and the murine T-ALL cell line S4924 and assayed the phosphorylation of downstream effectors. The serum-starved state was most informative for differentiating WT K-Ras from K-RasG12D and for testing the effects of the G37 and G64 site substitutions. Phosphorylated Akt (pAkt) levels were elevated in serum-deprived cells expressing K-RasD12/G37, whereas phosphorylated ERK (pERK) levels were normal (Figure 1B-C). In contrast, K-RasD12/G64 increased pERK, but not pAkt, levels (Figure 1B-C). K-RasD12/G64 expression in 3T3 fibroblasts induced morphological changes consistent with Raf/MEK/ERK pathway activation (Figure 1D).25
K-RasD12/G37 and K-RasD12/G64 induce T-ALL in vivo
We infected bone marrow cells from 5-fluorouracil-treated WT donor mice with a control MSCV-IRES-GFP vector or with viruses encoding WT Kras, KrasD12, KrasD12/G37, or KrasD12/G64 and transferred them into lethally irradiated WT mice. Recipients of cells transduced with the KrasD12 virus died early from hematopoietic failure (Figure 1E). Endogenous KrasD12 reduces the size of the hematopoietic stem cell compartment26 ; ectopic expression from the MSCV promoter likely exacerbates this defect, resulting in engraftment failure.
Mice transplanted with cells expressing K-RasD12/G37 or K-RasD12/G64 recovered hematologic function but began to die after 60 days (Figure 1E) from T-ALL characterized by elevated blood leukocyte counts, circulating GFP+ blasts, and thymic enlargement with invasion by CD4+/CD8+ blasts (Figure 1F-G and supplemental Figure 1B-C). Secondary recipients died of fulminant leukemia with a latency of 26.6 days (data not shown). The morphology and immunophenotype were identical in T-ALLs initiated by K-RasD12/G37 or K-RasD12/G64 expression, and Southern blot analysis revealed clonal retroviral integrations (supplemental Figure 1D). None of the mice transplanted with cells transduced with the MIG vector or with a virus encoding WT K-Ras developed hematologic disease.
Somatic NOTCH1 mutations are found in ∼50% of human T-ALLs27 and are also observed in mouse models of T-ALL characterized by endogenous KrasD12 expression.12,26,28 Western blot analysis of K-RasD12/G37 and K-RasD12/G64 leukemias with an antibody that detects activated (cleaved) Notch1 revealed abnormal fragments, and DNA sequencing confirmed Notch 1 PEST domain mutations in 7 of 11 primary T-ALLs that persisted in secondary recipients (supplemental Figure 1E and supplemental Table 2).
Ras and PTEN expression in KrasD12/G37 and KrasD12/G64-induced leukemias
We hypothesized that leukemias initiated by K-RasD12/G37 or K-RasD12/G64 might be under selective pressure to augment signaling through Ras effector pathways. To address this question, we biochemically assessed cell lines generated from 4 independent K-RasD12/G37 (E1-E4) and 6 independent K-RasD12/G64 (Y1-Y6) leukemias.
T-ALL cell lines expressing K-RasD12/G37 showed elevated Ras protein levels, particularly lines E2 and E4 (Figure 2A), which corresponded to elevated expression of K-RasD12/G37 (Figure 2B), and elevated viral Kras DNA copy number (Figure 2C). Three of 4 K-RasD12/G37 cell lines retained PTEN expression. Of these cell lines, 2 had low levels of pAkt and pERK, whereas cell line E3 showed a modest increase in pAkt under basal growth conditions (Figure 2A). We unexpectedly identified a somatic Pten mutation in cell line E4, which also had no detectable PTEN protein or mRNA expression and exhibited markedly elevated levels of pAkt (Figure 2A and supplemental Figure 2A-B). This mutation was also found in the T-ALL that gave rise to the cell line (supplemental Figure 2A).
Leukemias initiated by KrasD12/G37 and KrasD12/G64 expression demonstrate distinct signaling profiles. (A) Immunoblot of T-ALL cell lines under basal (B) and starved (S) conditions. Two control T-ALL cell lines with WT Kras (C) from a retroviral insertional mutagenesis screen11 were included. Ten cell lines were generated from independent leukemias induced by either KrasD12/G37 (E1-E4) or KrasD12/G64 (Y1-Y6). (B) Ras was immunoprecipitated from cell lines E1-E4 and then probed with an antibody that recognizes the D12 substitution. (C) quantitative PCR analysis of Kras expression in T-ALL cell lines compared with WT thymus.
Leukemias initiated by KrasD12/G37 and KrasD12/G64 expression demonstrate distinct signaling profiles. (A) Immunoblot of T-ALL cell lines under basal (B) and starved (S) conditions. Two control T-ALL cell lines with WT Kras (C) from a retroviral insertional mutagenesis screen11 were included. Ten cell lines were generated from independent leukemias induced by either KrasD12/G37 (E1-E4) or KrasD12/G64 (Y1-Y6). (B) Ras was immunoprecipitated from cell lines E1-E4 and then probed with an antibody that recognizes the D12 substitution. (C) quantitative PCR analysis of Kras expression in T-ALL cell lines compared with WT thymus.
In contrast, 5 of 6 K-RasD12/G64 T-ALL cell lines had normal Ras protein levels. PTEN was absent or barely detectable in all 5 cell lines, and reverse-transcription polymerase chain reaction (PCR) analysis revealed markedly reduced Pten mRNA levels, but no Pten mutations (Figure 2A, supplemental Figure 2B, and data not shown). K-RasD12/G64 T-ALL cells without PTEN expression had high basal pAkt levels that persisted during serum and cytokine deprivation (Figure 2A). These data support the idea that T-ALL cells overcome the deleterious effects of the Tyr-64 substitution by down-regulating PTEN expression, thereby activating PI3K signaling.
Acquired third site Kras mutations in KrasD12/G37 and KrasD12/G64 T-ALLs
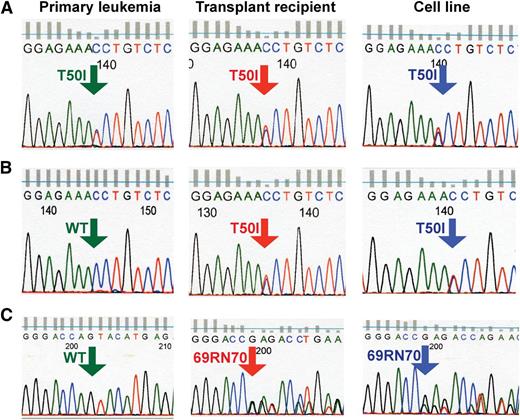
E1 and E3 had lower levels of Ras expression than E2 and E4 (Figure 2A), and DNA sequencing uncovered the same third site Kras mutation in both lines that introduced a threonine-to-isoleucine substitution at codon 50 (T50I) (Figure 3A-B). Germline T50I NRAS mutations cause Noonan syndrome, a developmental disorder characterized by hyperactive Raf/MEK/ERK signaling,29 and ectopic expression of K-RasT50I increased pERK levels.30 These data suggest that the acquired T50I mutation compensates for defective Raf binding in KrasD12/G37-induced leukemias. Y4 was distinct from the other K-RasD12/G64 lines, exhibiting elevated Ras levels and persistent PTEN expression (Figure 2A). DNA sequence analysis revealed Kras molecules containing the D12 and G64 substitutions along with a de novo in-frame insertion of an arginine and aspartic acid between codons 69 and 70 (69RN70) of the K-Ras switch II domain (Figure 3C).
Somatic third site Kras mutations in T-ALL. In cell lines E1 (A) and E3 (B), the single amino acid substitution T50I was present at ∼50% frequency based on relative abundance of sequence reads. It was detected at a similar frequency in the spleens of the secondary recipients used to generate these cell lines and in a single primary recipient. (C) The sequence GAGACC was inserted between amino acids 69 and 70 of Kras in T-ALL cell line Y4. This mutation is present in ∼50% of Kras transcripts in cell line Y4, at a lower frequency in the spleen of the secondary recipient used to generate this cell line, and is not seen in the primary leukemia.
Somatic third site Kras mutations in T-ALL. In cell lines E1 (A) and E3 (B), the single amino acid substitution T50I was present at ∼50% frequency based on relative abundance of sequence reads. It was detected at a similar frequency in the spleens of the secondary recipients used to generate these cell lines and in a single primary recipient. (C) The sequence GAGACC was inserted between amino acids 69 and 70 of Kras in T-ALL cell line Y4. This mutation is present in ∼50% of Kras transcripts in cell line Y4, at a lower frequency in the spleen of the secondary recipient used to generate this cell line, and is not seen in the primary leukemia.
To determine when each third site mutation occurred, we analyzed DNA from the recipients of MSCV-infected bone marrow, from secondary recipients, and from cell lines E1, E3, and Y4. Each cell line with a third site mutation also contained viral Kras DNA encoding the respective parental second site mutation, suggesting that the third site mutations arose de novo (supplemental Figure 3). Consistent with this idea, PCR-based sequencing of DNA uncovered the T50I mutation in primary T-ALL E1 and in a secondary recipient of T-ALL E3 (Figure 3A-B). The 69RN70 third site mutation was not detected in primary T-ALL Y4 but was present in the secondary recipient and further enriched in the cell line (Figure 3C). Together, these data suggest that third site Kras mutations are not required to initiate T-ALL but confer a strong clonal growth advantage.
K-Ras third site mutations restore full oncogenic activity
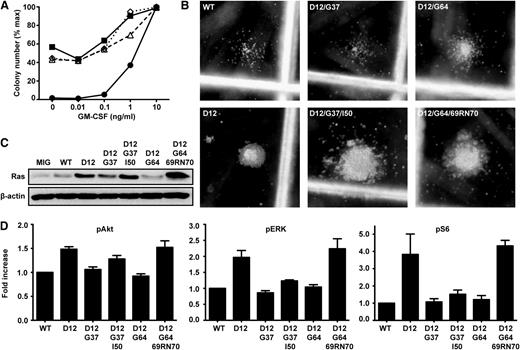
We assayed CFU-GM colony growth in transduced marrow cells to ask whether K-RasD12/G37/I50 and K-RasD12/G64/69RN70 are reactivated compared with the corresponding second site mutant proteins. Remarkably, progenitors expressing either third site mutation fully recapitulated the aberrant K-RasD12 growth phenotype, including cytokine-independent CFU-GM colony formation, pronounced GM-CSF hypersensitivity, and abnormal morphology (Figure 4A-B).
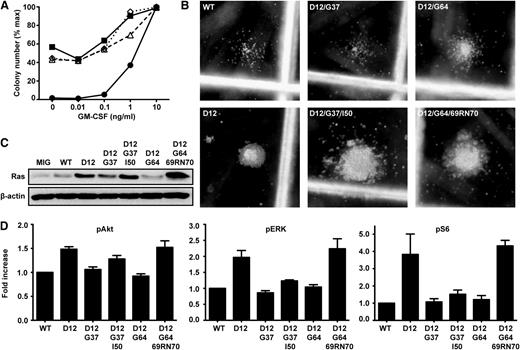
Acquired third site mutations restore oncogenic activity to KrasD12/G37 and KrasD12/G64. (A) CFU-GM formation of fetal liver cells expressing WT Kras (●), KrasD12 (▪), KrasD12/G37/I50 (Δ), or KrasD12/G64/69RN70 (♢). Data show the mean of 3 independent experiments. (B) Representative CFU-GM morphology from fetal liver cells expressing Kras mutant alleles grown in 0.1 ng/mL GM-CSF. (C) Ras expression in GFP+, Mac1+ fetal liver cells infected with MSCV viruses encoding different Kras alleles. (D) Levels of pERK, pAkt, and pS6 in GFP+, Mac1+ fetal liver cells infected with MSCV viruses encoding different Kras alleles, as determined by flow cytometry using phospho-specific antibodies. Phospho-protein levels in cells expressing WT K-Ras were set at 1 in each experiment. Data shown are derived from 6 independent experiments.
Acquired third site mutations restore oncogenic activity to KrasD12/G37 and KrasD12/G64. (A) CFU-GM formation of fetal liver cells expressing WT Kras (●), KrasD12 (▪), KrasD12/G37/I50 (Δ), or KrasD12/G64/69RN70 (♢). Data show the mean of 3 independent experiments. (B) Representative CFU-GM morphology from fetal liver cells expressing Kras mutant alleles grown in 0.1 ng/mL GM-CSF. (C) Ras expression in GFP+, Mac1+ fetal liver cells infected with MSCV viruses encoding different Kras alleles. (D) Levels of pERK, pAkt, and pS6 in GFP+, Mac1+ fetal liver cells infected with MSCV viruses encoding different Kras alleles, as determined by flow cytometry using phospho-specific antibodies. Phospho-protein levels in cells expressing WT K-Ras were set at 1 in each experiment. Data shown are derived from 6 independent experiments.
To assess the biochemical consequences of each third site mutation, we expressed K-RasD12/G37/I50 and K-RasD12/G64/69RN70 in mouse fetal hematopoietic cells. Cells expressing oncogenic K-RasD12 or either third site mutant proliferated vigorously and had higher Ras expression than GFP+ cells infected with MSCV vectors encoding WT K-Ras or either second site mutant (Figure 4C). K-RasD12/G37/I50 or K-RasD12/G64/69RN70 expression resulted in elevated pERK, pAkt, and pS6 levels compared with cells expressing WT K-Ras or the corresponding second site mutant proteins, which was particularly evident for K-RasD12/G64/69RN70 (Figure 4D and supplemental Figure 4A-B). The modest increase in pERK levels in hematopoietic cells expressing the T50I mutant protein is consistent with data from cell lines E1 and E3 (Figure 2A).
We examined the potential consequences of the T50I substitution and the 69RN70 insertion on predicted Ras-Raf and Ras-PI3K co-crystal structures. Although our analysis of T50I suggested potential effects on Ras dimer interactions, with the mutant protein reorienting the neighboring Ras molecule to enhance binding to Raf-1, we cannot exclude other potential consequences of this substitution (supplemental Figure 5A-B and supplemental Methods). The 69RN70 insertion is predicted to restore contact with PI3Kγ (supplemental Figure 5C).
Responses of T-ALL cells to chemical inhibitors
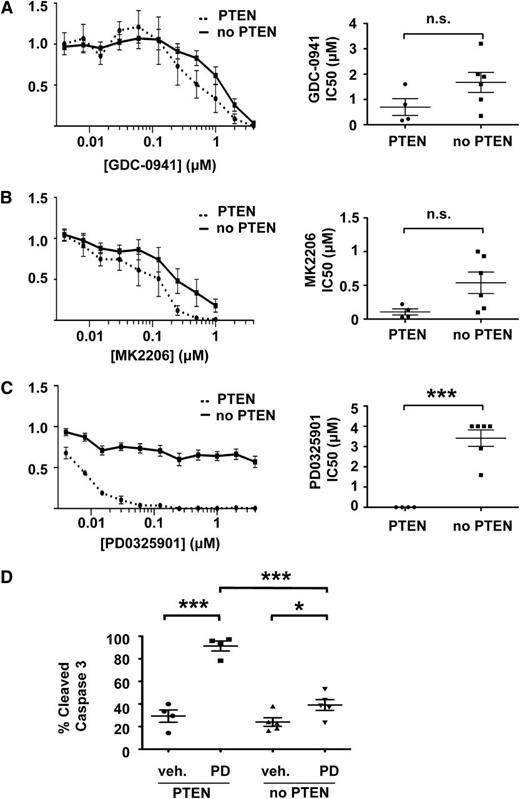
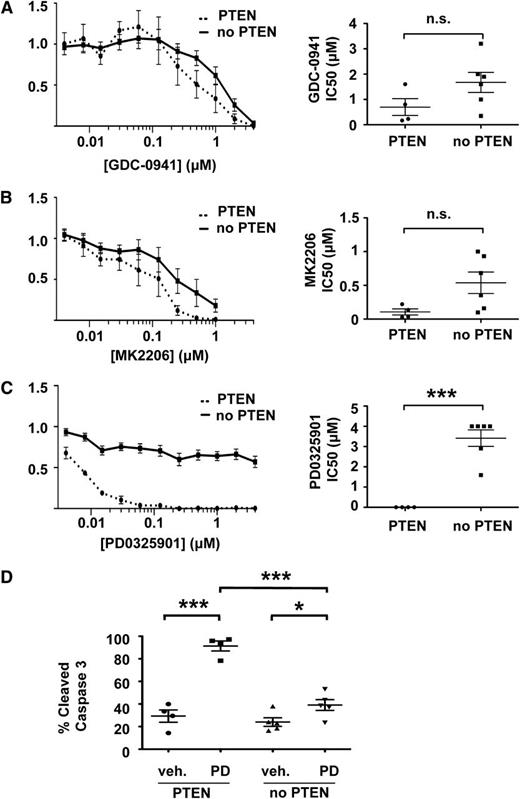
We exposed our T-ALL cell lines to inhibitors of PI3K (GDC-0941),31 Akt (MK2206),32 or MEK(PD0325901)33 to determine whether their biochemical characteristics and/or the presence of third site mutations correlated with drug sensitivity. GDC-0941 efficiently reduced pAkt levels in T-ALL cell lines (supplemental Figure 6A) and blocked their growth (Figure 5A). Although the IC50 was lower for lines that retained PTEN expression, this difference was modest (Figure 5A). Similarly, MK2206 both abrogated Akt phosphorylation and inhibited growth in a dose-dependent manner (Figure 5B and supplemental Figure 6B). As with GDC-0941, we observed similar IC50 values in T-ALL cell lines with and without intact PTEN that were treated with MK2206. Together, these data indicate that T-ALL cells are dependent on PI3K signaling for growth irrespective of basal pathway activation. The somewhat higher IC50 values observed in lines lacking PTEN expression likely reflect a requirement for greater drug concentrations to fully suppress pAkt.
PTEN loss confers resistance to inhibition of MEK but not PI3K or Akt. IC50s and mean growth curves of T-ALL cell lines with and without PTEN expression in varying doses of (A) the PI3K inhibitor GDC-0941, (B) the Akt inhibitor MK2206, and (C) the MEK inhibitor PD0325901. (D) PD0325901 induces apoptosis in PTEN-positive, but not PTEN-negative, cell lines. Curves indicate mean growth of 4 PTEN-positive cell lines (E1, E2, E3, and Y4) and 6 PTEN-negative cell lines (E4, Y1, Y2, Y3, Y5, and Y6) ± SEM. ***P < .001, *P < .05.
PTEN loss confers resistance to inhibition of MEK but not PI3K or Akt. IC50s and mean growth curves of T-ALL cell lines with and without PTEN expression in varying doses of (A) the PI3K inhibitor GDC-0941, (B) the Akt inhibitor MK2206, and (C) the MEK inhibitor PD0325901. (D) PD0325901 induces apoptosis in PTEN-positive, but not PTEN-negative, cell lines. Curves indicate mean growth of 4 PTEN-positive cell lines (E1, E2, E3, and Y4) and 6 PTEN-negative cell lines (E4, Y1, Y2, Y3, Y5, and Y6) ± SEM. ***P < .001, *P < .05.
We next assessed the effects of blocking Raf/MEK/ERK signaling. Remarkably, loss of PTEN expression and elevated pAkt levels strongly correlated with resistance to PD0325901, despite equivalent target inhibition (Figure 5C and supplemental Figure 6C). The median IC50 values were 0.00625 µM and more than 4 µM in PTEN-positive and PTEN-negative cells, respectively (Figure 5C). Resistance to PD0325901 in PTEN-negative cells appears to occur via abrogation of apoptosis (Figure 5D).
T-ALL cell lines with PI3K pathway activation and human ETP ALL have similar gene expression profiles
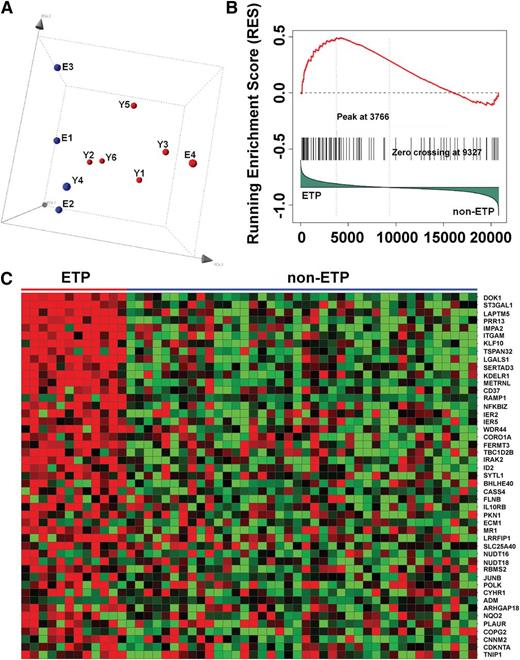
Microarray-based gene expression profiling on our T-ALL cell lines revealed a single cluster of cell lines without PI3K pathway activation (E1, E2, and Y4) and a second cluster of cell lines with loss of PTEN expression and markedly elevated pAkt levels (Figure 6A). E3, the K-RasD12/G37 line with a modest increase in basal pAkt (Figure 2A), did not clearly segregate with either group (Figure 6A). Gene set enrichment analysis showed that cell lines with PI3K pathway activation demonstrated differential expression of many genes functionally linked to JAK/STAT and Raf/MEK/ERK signaling (supplemental Tables 3-8). The expression profile of these T-ALL cell lines is highly similar to that of ETP T-ALL, an aggressive cancer characterized by a high risk for treatment failure and frequent RAS mutations (Figure 6B-C).34,35 This association persisted when the data were reanalyzed to include E4 with either group of T-ALL cell lines (supplemental Figure 6A-B).
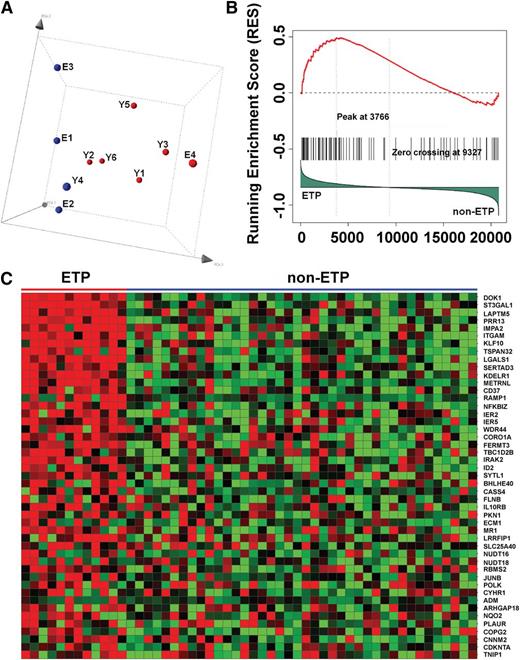
PI3K-activated T-ALL cell lines and human ETP ALLs have similar gene expression profiles. (A) Principal component analysis of the gene expression profiling data of all 10 mouse T-ALL cell lines using 200 representative genes selected by k-means algorithm, showing cases clustered according to PI3K activation status (red, activated; blue, not activated). (B) Gene set enrichment analysis demonstrates significant enrichment of the top 100 mouse PI3K up-regulated genes in ETP ALL (P = .057; false discovery rate, 0.224). (C) Heat map of the leading-edge mouse PI3K up-regulated genes in gene set enrichment analysis, showing overexpression of these genes in ETP ALL.
PI3K-activated T-ALL cell lines and human ETP ALLs have similar gene expression profiles. (A) Principal component analysis of the gene expression profiling data of all 10 mouse T-ALL cell lines using 200 representative genes selected by k-means algorithm, showing cases clustered according to PI3K activation status (red, activated; blue, not activated). (B) Gene set enrichment analysis demonstrates significant enrichment of the top 100 mouse PI3K up-regulated genes in ETP ALL (P = .057; false discovery rate, 0.224). (C) Heat map of the leading-edge mouse PI3K up-regulated genes in gene set enrichment analysis, showing overexpression of these genes in ETP ALL.
Discussion
CFU-GM progenitors expressing second site K-RasD12 mutant proteins that only signal through Raf/MEK/ERK (K-RasD12/E38) or PI3K/Akt (K-RasD12/C40) displayed normal growth in methylcellulose cultures over a range of GM-CSF doses (Figure 1A). In contrast, Kras oncogenes encoding proteins that retain the ability to engage Ral-GDS but are defective for either PI3K/Akt (K-RasD12/G64) or Raf/MEK/ERK (K-RasD12/G37) pathway activation demonstrated in vitro and in vivo transforming activity. The leukemias initiated by these second site mutant alleles underwent clonal evolution in vivo, characterized by rapid outgrowth of cells with secondary mutations that overcome impaired signaling properties. Although the use of a retroviral transduction/transplantation system likely facilitated transformation in our studies, Ras protein levels were not elevated in most K-RasD12/G64 cell lines (Figure 2A). The absence of hematologic cancers in mice transplanted with cells overexpressing WT Kras from the same MSCV vector further demonstrates that K-RasD12/G37 and K-RasD12/G64 have “gain of function” oncogenic activity.
Endogenous KrasD12 expression causes both T-ALL and myeloid leukemia in mice, which is influenced by a number of factors.26,28,36-39 Use of the Mx1-Cre transgene to express a latent KrasD12 oncogene causes an aggressive myeloproliferative neoplasm,36,39 but transferring bone marrow from Mx1-Cre; KrasD12 mice into irradiated recipients induces T-ALL,26,28,38 and NRAS, KRAS, and other genes commonly mutated in myeloid malignancies are also mutated in ETP T-ALL.35 Although the signaling profiles of these aggressive cancers and their susceptibility to targeted therapies have not been reported, our data linking gene expression in ETP-ALL to aberrant PI3K signaling and the observation that T-ALL cell lines with Kras mutations are sensitive to PI3K inhibition12 supports developing clinical trials in which PI3K pathway inhibitors are administered with conventional antileukemia therapy. The paradigm of combining targeted and conventional agents has recently been applied successfully to BCR-ABL-positive ALL.40
PI3K binds to and is activated by Ras-GTP6,19,41 ; however, the precise role of PI3K signaling in Ras-induced cancer initiation and maintenance is uncertain. An elegant study by Gutpa et al23 demonstrating dramatic inhibition of KrasD12-induced lung cancer in mice expressing a knock-in PI3K p110α protein that is defective for Ras binding supports an essential role of PI3K activation in tumorigenesis.42 In contrast, PI3K/Akt-mediated growth and survival of colon cancer cell lines appears to be dependent on receptor tyrosine kinase signaling, but independent of oncogenic KRAS.43 Genetic analysis of T-ALL specimens implicated Ras/PI3K pathway mutations as important drivers of leukemic growth.9 Consistent with this idea, T-ALLs initiated by Kras oncogenes with second site mutations frequently silenced PTEN and activated Akt, demonstrating strong selective pressure for leukemia cells to acquire aberrant PI3K/Akt signaling in vivo.
The role of aberrant Raf/MEK/ERK signaling in T-ALL is less clear. The identification of somatic third site T50I mutations that enhance signaling and fully transform progenitor growth supports the idea that the Raf/MEK/ERK cascade contributes to leukemogenesis. Indeed, T-ALL cell lines that retained PTEN expression were susceptible to the MEK inhibitor PD0325901. However, our studies suggest that PI3K pathway activation confers resistance by providing a survival signal in T-ALL cells that overcomes the proapoptotic effects of MEK inhibition. This observation is supported by in vivo data showing that administering a MEK inhibitor to Mx1-Cre; KrasD12 mice abrogated signs of myeloproliferative neoplasm, but that some animals nevertheless progressed to T-ALL during treatment.44
The discoveries of Pten inactivation in a K-RasD12/G37 T-ALL and a 69RN70 insertion with elevated pERK levels in a K-RasD12/G64 leukemia indicate unexpected plasticity in the spectrum of secondary mutations and suggest that enhanced signaling through an already-activated pathway can compensate for impaired binding to another effector. This idea is consistent with recent studies of resistance to MEK inhibitors showing that the underlying mechanisms include amplification of KRAS and BRAF oncogenes45 and overexpression of RasGRP guanine nucleotide exchange factors.46
Although the Ras GTPase switch is widely viewed as “undruggable,”47 the development of chemical inhibitors of other Ras domains is an emerging area of investigation.4 As strategies for targeting oncogenic Ras are tested in advanced cancers, acquired resistance is inevitable. Indeed, our data demonstrate that KrasD12 can evolve rapidly in vivo by acquiring novel secondary mutations, suggesting that “on target” mutations leading to drug resistance will emerge in patients who are treated with inhibitors of oncogenic Ras. Indeed, the recent identification of insertions similar to the 69RN70 alteration in human lung and colorectal cancers48,49 underscores the relevance of this potential mechanism in facilitating malignant growth.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Judith Leopold (Pfizer, Inc.) for PD0325901, to Deepak Sampath (Genentech) for GDC-0941, and to D. Tuveson, P. Perez-Mancera, S. Lowe, and B. Braun for discussion and advice.
This work was supported by National Institutes of Health grants R37CA72614, U54CA143874, and T32CA09043; by a Specialized Center of Research award from the Leukemia and Lymphoma Society (7019-04); by the Frank A. Campini Foundation; and by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Authorship
Contribution: A.S. and A.F.W. generated reagents, designed experiments, performed research, analyzed data, and wrote the manuscript; K.L.D. designed experiments, performed research, and analyzed data; E.R.H.-T. performed research; J.X. generated reagents and performed experiments; C.G.M. and C.Z. performed research, analyzed data, and assisted in writing and editing the manuscript; S.-C.C. and X.S. performed bioinformatic and statistical analysis of data and assisted in writing and editing the manuscript; J.R.D. analyzed and verified data and assisted in writing and editing the manuscript; G.E.B. performed research, analyzed data, and assisted in writing and editing the manuscript; and K.M.S. provided overall oversight of the project, designed and supervised the experiments, reviewed, verified, and analyzed data, established collaborations, and wrote the manuscript.
Conflict-of-interest disclosure: G.E.B. and C.Z. are employees of Plexxikon and hold equity in the company. K.M.S. is an American Cancer Society Research Professor, C.G.M. is a Pew Scholar in the Biomedical Sciences, and J.X. is an American Cancer Society Fellow. The remaining authors declare no competing financial interests.
Correspondence: Kevin M. Shannon, Helen Diller Family Cancer Research Building, University of California, San Francisco, 1450 3rd St; Room 240, San Francisco, CA 94158-9001; e-mail: shannonk@peds.ucsf.edu.
References
Author notes
A.S. and A.F.W. contributed equally to this study.