Key Points
Telomere length in MCL is variable but does not correlate with disease characteristics and survival.
Abstract
Telomere shortening is of pathogenic and prognostic importance in cancers. In the present study, we analyzed telomere length in 73 mantle cell lymphoma (MCL), 55 chronic lymphocytic leukemia (CLL), and 20 normal B-cell samples using quantitative PCR (Q-PCR) to study its association with disease characteristics and outcome. Telomere length was found to be highly variable in MCL (range, 2.2-13.8 kb; median, 4.3 kb). Telomere dysfunction in MCL was evident from comparison with normal B cells (median, 7.5 kb), but had no significant association with any biologic or clinical feature. This was in contrast to CLL, in which a significant correlation of short telomeres with poor prognostic subgroups was confirmed. There was a trend toward an increased number of genomic aberrations with shortening of telomeres in MCL. No difference in survival was observed between the groups with short and long telomeres, indicating that, as opposed to CLL, telomere length is not of prognostic relevance in MCL.
Introduction
Mantle cell lymphoma (MCL) is a specific subtype of non-Hodgkin lymphoma with a median overall survival of approximately 4-5 years and significant heterogeneity.1 Several studies have identified various pathogenic and prognostic factors in MCL ranging from genomic aberrations2,3 ; IGHV mutation status4,5 ; gene mutations in ATM,6 TP53,7 CCND1,8 and NOTCH19 ; proliferation index (Ki67)10 ; and expression levels of genes11,12 to the establishment of MCL international prognostic index (MIPI) based on age, performance status, lactate dehydrogenase, and leukocyte count at diagnosis.13 MCL shows a relatively high number of genomic alterations per case2,3 and one of the many causes for genomic instability is telomere dysfunction.
Short, dysfunctional telomeres have been associated with chromosomal end-to-end fusions that predispose the cells to chromosomal aberrations, genomic complexity, and malignant transformation.14 In cancers, there may be a relation between telomere dysfunction and an increase in genomic complexity, clonal evolution, and, consequently, to chemoresistance and poor survival.
In CLL, telomere length is highly variable and short telomeres have been associated with unmutated IGHV status, high-risk genomic aberrations, and poor outcome in univariate and multivariate analyses.15-17 In contrast, study of pathogenic and prognostic role of telomere dysfunction in MCL using a well-characterized, representative cohort has not been done. Earlier studies on telomere length in MCL were carried out with a limited number of samples18,19 and analyses of telomere length associations with other biologic disease characteristics and prognostic factors were not performed.19,20 In the present study, we analyzed telomere length in 73 well-characterized MCL samples from lymph nodes (n = 51) and peripheral blood (n = 22) to evaluate associations with biologic and clinical disease characteristics and outcome.
Methods
Tumor samples of 73 MCL and 55 CLL patients treated at the Universities of Ulm, Würzburg, and Heidelberg were obtained after informed consent in accordance with the Declaration of Helsinki and approval from the institutional review boards of each institution. DNA from lymph node samples was isolated from fresh/frozen tissue blocks and that from peripheral blood samples was isolated from PBMCs. Peripheral blood samples from older healthy donors (median age, 56 years; range, 46-68 years) were obtained for comparison of telomere length of MCL with normal B cells and DNA was isolated from CD19-sorted PBMCs. Clinical and molecular characteristics of the MCL and CLL cohorts used for the study are summarized in Table 1. Analyses of genomic aberrations by FISH, proliferation indices by Ki67 stain, array CGH, and sequencing of IGHV and TP53 were done as described previously.10,21,22 Measurement of telomere length was carried out using a Q-PCR–based technique.23 The primers used to amplify telomere and single-copy genes (SCG) were tel1b and tel2b and HBG3 and HBG4, respectively.16,24 Synthetic oligonucleotide standards for telomere (84 bp) and SCG (81 bp) PCR were used to quantify the amount of telomere sequence per sample. Twenty-five nanograms of DNA was used per reaction in triplicates for the telomere and SCG PCRs. Three telomere length controls were included in every plate to detect plate-to-plate variations. The Q-PCR technique was validated by terminal restriction fragment length (TRF) analysis25 and a correlation of R2 = 0.8516 was obtained (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The TRF value of telomere length for each sample was calculated from the linear regression of Q-PCR versus TRF. Tumor cell content of the samples was estimated by the percentage of cells harboring the most prevalent genomic aberrations as analyzed by FISH (n = 57). Samples with no (n = 10) or partial (7 of 57) FISH data were analyzed for relative expression of CD3 using Q-PCR to control for sample purity (median: 0.15) as described previously.4 Because the MCL samples used in the study were in general associated with high tumor load, 6 samples with no FISH or CD3 expression were also included for analysis. The lymph node and peripheral blood MCL samples were analyzed separately because of potential differences in telomere lengths between tissues. We included the CLL cohort and B cells from healthy donors as control groups for comparison with MCL.
Results and discussion
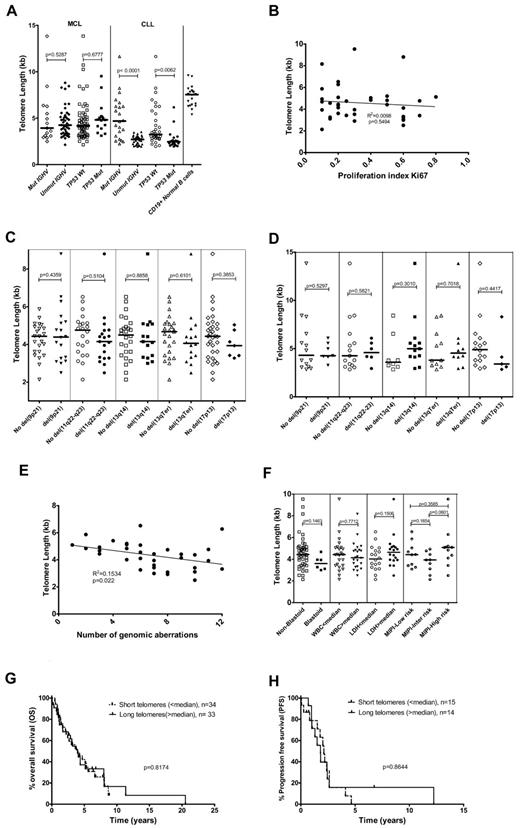
Telomere lengths of MCL and CLL samples were significantly shorter than those of normal B cells, indicating the occurrence of telomere dysfunction in MCL and CLL (Figure 1A). Telomere length has been associated with IGHV mutation status in CLL in several studies.15-17 In the current series, the association of short telomeres with an unmutated IGHV status was confirmed in CLL samples (P < .0001), whereas in the MCL cohort, no significant correlation was found (Figure 1A). In addition, there was no association of telomere length with the presence or absence of TP53 mutations in MCL cases, whereas CLL cases with a TP53 mutation showed a significant association with short telomeres (Figure 1A). There were no significant differences in telomere length comparing MCL samples from the lymph nodes and peripheral blood (supplemental Figure 2).
Correlation of telomere length with biologic and clinical factors. (A) Telomere length distribution in MCL and CLL samples with mutated/unmutated IGHV and mutant/wild-type TP53 and in normal B cells from healthy donors. (B) Correlation of telomere length with proliferation index (Ki67) levels. (C) Telomere length distribution among MCL lymph node samples with and without different genomic aberrations such as del(9p21), del(11q22-q23), del(13q14), del(13qTerminal), and del(17p13). (D) Telomere length distribution among MCL peripheral blood samples with and without different genomic aberrations such as del(9p21), del(11q22-q23), del(13q14), del(13qTerminal), and del(17p13). (E) Telomere length variation with the number of genomic aberrations detected using FISH and/or array-CGH (2 outliers with 0 and 1 genomic aberration detected were removed). (F) Relation of telomere lengths to clinical features in MCL samples. (G) Overall survival in the short (< median) and long telomere (> median) subgroups (median, 48 months; P = .8174). (H) Progression-free survival in the short (< median) and long telomere (> median) subgroups (median 25 vs 22 months; P = .8644). The lines in panels A, C, D, and F represent the median.
Correlation of telomere length with biologic and clinical factors. (A) Telomere length distribution in MCL and CLL samples with mutated/unmutated IGHV and mutant/wild-type TP53 and in normal B cells from healthy donors. (B) Correlation of telomere length with proliferation index (Ki67) levels. (C) Telomere length distribution among MCL lymph node samples with and without different genomic aberrations such as del(9p21), del(11q22-q23), del(13q14), del(13qTerminal), and del(17p13). (D) Telomere length distribution among MCL peripheral blood samples with and without different genomic aberrations such as del(9p21), del(11q22-q23), del(13q14), del(13qTerminal), and del(17p13). (E) Telomere length variation with the number of genomic aberrations detected using FISH and/or array-CGH (2 outliers with 0 and 1 genomic aberration detected were removed). (F) Relation of telomere lengths to clinical features in MCL samples. (G) Overall survival in the short (< median) and long telomere (> median) subgroups (median, 48 months; P = .8174). (H) Progression-free survival in the short (< median) and long telomere (> median) subgroups (median 25 vs 22 months; P = .8644). The lines in panels A, C, D, and F represent the median.
In somatic cells, the main mechanism of telomere shortening is attributed to the end replication problem during DNA replication, which therefore should be associated with the cellular proliferation index. However, we observed no correlation between telomere length and the proliferation index in the MCL samples (Figure 1B). This might be because of variable telomerase activity among the different MCL subsets. MCL cases of the blastoid subtype showed a tendency to have shorter telomeres, but the difference was not significant (Figure 1F).
Telomere lengths of the MCL samples did not show significant association with specific genomic aberrations. Only slight variations in median telomere length were observed among the MCL subgroups with different genomic aberrations in the lymph node (Figure 1C) and peripheral blood samples (Figure 1D). In contrast, in the CLL samples, a significant association of short telomeres with the high-risk aberrations del(17p13) and del(11q22-q23) was observed (supplemental Figure 3).16,17
Because telomere shortening could contribute to genomic instability and therefore to an increased incidence of genomic alterations, we analyzed the association of telomere length with the number of genomic aberrations. A weak but significant correlation (R2 = 0.1534, P = .022) was observed comparing telomere length with the number of genomic aberrations (Figure 1E). However, analysis of the relation of telomere length with clinical features showed no correlation with lactate dehydrogenase levels, WBC, or with MIPI groups (Figure 1F and supplemental Figure 4).
In the present study, we analyzed the impact of telomere length on survival in MCL cases by categorizing the patients into short (< median) and long (> median) telomere subgroups. No significant difference in overall survival (median, 48 months; P = .8174; Figure 1G) or progression-free survival (median, 25 vs 22 months; P = .8644; Figure 1H) was identified between the 2 subgroups. In contrast, when evaluating the impact of established prognostic factors on outcome to verify that our cohort was representative, we observed the expected correlations of Ki67 (P = .0001), MIPI index (P = .007), and B symptoms (P = .0119) with disease outcome (data not shown).
In summary, the comprehensive characterization of MCL cases using a large cohort with mature follow-up showed that telomeres in MCL samples were possibly dysfunctional by comparison with normal B cells but were not associated with any of the established clinical or biologic factors relevant for pathogenesis or prognostication and disease outcome. This is in marked contrast to CLL, in which telomere length has been associated with biologic disease characteristics, pathogenic mechanisms, and survival in multivariate analysis.15-17
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sabrina Kless for excellent technical assistance.
This work was supported by funding from Deutsche Forschungsgemeinschaft (SFB1074, subproject B1) and the European Mantle Cell Lymphoma Network. B.M.C.J. was supported by a fellowship from the International Graduate School in Molecular Medicine, Ulm, Germany.
Authorship
Contribution: B.M.C.J. performed the experiments, interpreted the data, and wrote the manuscript; D.K. interpreted the data, edited the manuscript, and provided vital material; A.L. and D.M. interpreted the data and edited the manuscript; M.H. performed the experiments; G.O., A.R., T.F.E.B., and P.M. interpreted the data, edited the manuscript, and provided vital material; T.Z. designed the research and conceived the study; T.Z. and H.D. interpreted the data and edited the manuscript; and S.S. conceived the study, designed the research, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephan Stilgenbauer, MD, Department of Internal Medicine III, University of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: stephan.stilgenbauer@uniklinik-ulm.de.