Key Points
Bif-1 acts as a haploinsufficient tumor suppressor in Myc-induced lymphomagenesis.
Bif-1 plays a key role in mitophagy to maintain chromosome stability.
Abstract
Malignant transformation by oncogenes requires additional genetic/epigenetic changes to overcome enhanced susceptibility to apoptosis. In the present study, we report that Bif-1 (Sh3glb1), a gene encoding a membrane curvature–driving endophilin protein, is a haploinsufficient tumor suppressor that plays a key role in the prevention of chromosomal instability and suppresses the acquisition of apoptosis resistance during Myc-driven lymphomagenesis. Although a large portion of Bif-1–deficient mice harboring an Eμ-Myc transgene displayed embryonic lethality, allelic loss of Bif-1 dramatically accelerated the onset of Myc-induced lymphoma. At the premalignant stage, hemizygous deletion of Bif-1 resulted in an increase in mitochondrial mass, accumulation of DNA damage, and up-regulation of the antiapoptotic protein Mcl-1. Consistently, allelic loss of Bif-1 suppressed the activation of caspase-3 in Myc-induced lymphoma cells. Moreover, we found that Bif-1 is indispensable for the autophagy-dependent clearance of damaged mitochondria (mitophagy), because loss of Bif-1 resulted in the accumulation of endoplasmic reticulum–associated immature autophagosomes and suppressed the maturation of autophagosomes. The results of the present study indicate that Bif-1 haploinsufficiency attenuates mitophagy and results in the promotion of chromosomal instability, which enables tumor cells to efficiently bypass the oncogenic/metabolic pressures for apoptosis.
Introduction
Tumorigenesis is a multistep process by which normal cells progressively accumulate genetic mutations and epigenetic modifications that allow for sustained cell proliferation. During this process, overproliferated precancerous cells are exposed to a variety of environmental stresses (eg, growth factor limitation and nutrient starvation) that induce apoptosis. Evasion of apoptosis is therefore widely acknowledged to be a critical step in tumor development and a hallmark of most types of cancers.1
The Bcl-2 family of proteins, which is composed of antiapoptotic (eg, Bcl-2, Bcl-xL, and Mcl-1), multidomain proapoptotic (eg, Bax and Bak), and BH3-only proapoptotic regulatory members, plays a central role in the modulation of outer mitochondrial membrane (OMM) permeabilization to regulate the activation of downstream caspases and apoptosis. Overexpression of the antiapoptotic Bcl-2 family members or deletion of the proapoptotic members has been shown to synergize oncogene-driven tumorigenesis.2 Moreover, recent high-resolution somatic copy-number alterations analyses across multiple cancer types have revealed that human cancer cells frequently amplify Bcl-xL- and Mcl-1–containing regions and depend on the expression of these antiapoptotic genes for survival.3
Macroautophagy, hereafter referred as autophagy, is a lysosome-mediated intracellular degradation process for turnover of proteins and organelles. It has been shown that inhibition of autophagy by a lysosomal inhibitor enhances p53-mediated apoptosis and tumor regression in a Myc oncogene–induced mouse lymphoma model.4 Moreover, autophagy facilitates cancer cell survival by maintaining functional mitochondria to support tumor development during oncogenic Ras activation.5 Elevated levels of autophagy were also observed in tumor xenograft models,6,7 supporting the notion that autophagy functions to promote tumor survival in response to exposure to metabolic stresses. Paradoxically, however, the induction of autophagy is generally suppressed or limited in many tumor tissues through mechanisms such as the haploid deletion of the autophagy-related gene Beclin 1 or constitutive activation of the PI3K/MTOR signaling pathway.8 Moreover, allelic loss of Beclin 1 has been shown to promote spontaneous or Myc oncogene–induced tumorigenesis in mice.9,10 Furthermore, inhibition of autophagy in Bcl-2–overexpressing, apoptosis-incompetent cells has been shown to result in the accumulation of polyubiquitinated proteins and damaged mitochondria that promote the production of reactive oxygen species and chromosomal instability.11,12 Therefore, although the prosurvival function of autophagy for cancer cell growth often overwhelms the tumorigenic effects of autophagy deficiency,9 defects in autophagy in certain circumstances may promote tumorigenesis.
Bif-1 (SH3GLB1/Endophilin B1), a member of the membrane curvature–driving endophilin family of proteins, regulates the induction of apoptosis and autophagy.13 In humans, Bif-1 is located on chromosome 1p22, a region that is frequently deleted in neoplasms.14 Down-regulation of Bif-1 expression has been reported in a variety of cancers.15-19 We reported previously that knockdown of Bif-1 enhances the tumorigenicity of HeLa cells in vivo20 and the targeted deletion of Bif-1 promotes spontaneous tumor development in mice.21 Although these studies highlight the importance of Bif-1 as a tumor suppressor, the mechanism behind the promotion of tumorigenesis by Bif-1 deficiency remains unclear. In the present study, we provide evidence that Bif-1 regulates autophagy-dependent mitochondrial turnover and acts as a haploinsufficient tumor suppressor to prevent chromosomal instability and the acquisition of apoptosis resistance during Myc-driven lymphomagenesis.
Methods
Mice
C57BL/6J wild-type mice and B6.Cg-Tg(IghMyc)22Bri/J (Eμ-Myc) mice were purchased from The Jackson Laboratory. All animal protocols were approved by the Pennsylvania State University Animal Care and Use Committee. B6.Bif-1+/− and B6.Bif-1−/− female mice were generated by backcrossing B6/129.Bif-1−/− mice20 with C57BL/6J wild-type mice for 12 generations and intercrossed with Eμ-Myc transgenic male mice. PCR-based genotyping is described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry and karyotype analyses
Splenocytes were prepared from prelymphomatous mice. The cells were stained with 100nM MitoTracker Green FM (M-7514; Invitrogen) for 30 minutes and analyzed using an LSR II Flow Cytometer System (BD Biosciences) and FlowJo Version 9.3.2 software (TreeStar). For karyotype analyses, the cells were treated with 5 μg/mL of lipopolysaccharide overnight to stimulate cell-cycle progression. Metaphase spreads were then prepared using standard procedures after 10 μg/mL of Colcemid (KaryoMax, 15212-012; Invitrogen) treatment for 2 hours. Each metaphase was evaluated for chromosomal aneuploidy. The total number of chromosomes in each metaphase was counted from photographs.
Electron microscopy
Cells seeded on Thermanox plastic coverslips (72280; Electron Microscopy Sciences) were treated with 30μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) or DMSO for 24 hours and then fixed in Karnovsky fixative (1.5% paraformaldehyde/3% glutaraldehyde in 0.1M cacodylate buffer, pH 7.3) containing 4.2mM ethyleneglycol-bis(b-aminoethyl)-N,N,N′,N′-tetracetic acid for 30 minutes at room temperature, followed by 2 hours at 4°C. The cells were postfixed in 1% osmium tetroxide/1.5% potassium ferrocyanide in 0.1M sodium cacodylate, pH 7.3, for 30 minutes at 4°C, soaked in 1% uranyl acetate for 2 hours, dehydrated in a graded series of ethanol, embedded in EMbed 812 resin (14120; Electron Microscopy Sciences), and sectioned at a thickness of 70-90 nm. The sections were mounted on mesh copper grids, stained with aqueous uranyl acetate and lead citrate, and analyzed using a JEM 1400 transmission electron microscope (JEOL).
Results
Bif-1 haploinsufficiency accelerates Myc-driven lymphomagenesis
To further explore the potential of Bif-1 as a tumor suppressor, Bif-1+/− mice with a C57BL/6J genetic background were crossed with Eμ-Myc transgenic mice, a well-established model for mouse B-cell lymphoma.22 We found that knockout of Bif-1 accelerated Eμ-Myc–induced tumor onset and mortality compared with Eμ-Myc/Bif-1+/+ mice (median onset, 65 vs 107 days, respectively, P = .0006; Figure 1A). Eμ-Myc/Bif-1+/− mice also exhibited a shortened life span compared with Eμ-Myc/Bif-1+/+ mice (median onset, 75 days, P < .0001). Although 36.5% of Eμ-Myc/Bif-1+/+ mice (19 of 52) survived more than 135 days, none of the Eμ-Myc/Bif-1+/− (0 of 53) or Eμ-Myc/Bif-1−/− (0 of 9) mice did. No statistically significant difference in disease onset or mortality was observed between Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− mice. Eμ-Myc/Bif-1+/+ and Eμ-Myc/Bif-1+/− mice were born at the expected Mendelian ratio and appeared healthy at birth (data not shown). Similarly, newborn Eμ-Myc/Bif-1−/− mice appeared normal and indistinguishable from their wild-type littermates. However, although Bif-1−/− mice without the Eμ-Myc transgene were born at normal Mendelian ratios,20 the frequency of birth of Eμ-Myc/Bif-1−/− mice was significantly lower than expected (eg, 9.8% vs 25.0% in Eμ-Myc/Bif-1+/− and Bif-1−/− matings; supplemental Table 1). To determine whether a portion of Eμ-Myc/Bif-1−/− mice die during embryogenesis, embryos were dissected at several developmental stages. We found few Eμ-Myc/Bif-1−/− embryos with no apparent developmental defects in addition to several resorbed embryos at embryonic days 12.5 (E12.5) and E14.0 (supplemental Figure 1 and data not shown). A higher proportion of Eμ-Myc/Bif-1−/− embryos were detected at E9.5, although 2 of 5 isolated deciduae already contained developmentally arrested or resorbed Eμ-Myc/Bif-1−/− embryos. Therefore, we conclude that the majority of Eμ-Myc/Bif-1−/− mice die during embryogenesis, presumably around E9.5. These results indicate that not only does the loss of Bif-1 promote embryonic lethality in Eμ-Myc transgenic mice, but Bif-1 also functions as a haploinsufficient tumor suppressor gene in Eμ-Myc–induced lymphomagenesis.
Loss of Bif-1 promotes Myc-driven lymphoma development. (A) Kaplan-Meier tumor-free survival of Eμ-Myc/Bif-1+/+ (n = 52; median onset, 107 days), Eμ-Myc/Bif-1+/− (n = 53; median onset, 75 days), and Eμ-Myc/Bif-1−/− (n = 9; median onset, 65 days) mice is shown. Statistical significance was determined using a log-rank test. (B) Bif-1 haploinsufficiency enhances the frequency of thymic lymphoma in Eμ-Myc–transgenic mice. Gross images of typical lymphomatous Eμ-Myc/Bif-1+/+ and Eμ-Myc/Bif-1+/− mice with thymic lymphoma are shown. The lymph nodes and thymus are outlined by yellow circles and a white circle/arrow, respectively. Statistical significance was determined using a Fisher exact test. (C) The weights of spleens from all of the mice used in this figure were measured at the onset of lymphoma and are shown as a box plot. The lines, boxes, and error bars represent median values, 25-75 percentiles, and the 10-90 percentiles, respectively. (D-E) Allelic loss of Bif-1 suppresses lung perivascular tumor formation in Eμ-Myc–transgenic mice. The lungs (D) and livers (data not shown) from lymphomatous Eμ-Myc/Bif-1+/+ (n = 34; ii), Eμ-Myc/Bif-1+/− (n = 38; iii), Eμ-Myc/Bif-1−/− (n = 8; iv), and control Bif-1+/+ mice (i) were stained with H&E. The degree of lymphoblast infiltration to pulmonary alveolar septa (D) or hepatic sinusoids (E) and perivascular interstitial stroma expanded by lymphoblasts (D-E) were scored as follows: 0 = normal; 1 = < 5%; 2 = 6%-15%; 3 = 16%-40%; and 4 = > 40%. The lines represent median values. Statistical significance was determined using a 2-way ANOVA followed by a Scheffe posthoc test. *P < .05.
Loss of Bif-1 promotes Myc-driven lymphoma development. (A) Kaplan-Meier tumor-free survival of Eμ-Myc/Bif-1+/+ (n = 52; median onset, 107 days), Eμ-Myc/Bif-1+/− (n = 53; median onset, 75 days), and Eμ-Myc/Bif-1−/− (n = 9; median onset, 65 days) mice is shown. Statistical significance was determined using a log-rank test. (B) Bif-1 haploinsufficiency enhances the frequency of thymic lymphoma in Eμ-Myc–transgenic mice. Gross images of typical lymphomatous Eμ-Myc/Bif-1+/+ and Eμ-Myc/Bif-1+/− mice with thymic lymphoma are shown. The lymph nodes and thymus are outlined by yellow circles and a white circle/arrow, respectively. Statistical significance was determined using a Fisher exact test. (C) The weights of spleens from all of the mice used in this figure were measured at the onset of lymphoma and are shown as a box plot. The lines, boxes, and error bars represent median values, 25-75 percentiles, and the 10-90 percentiles, respectively. (D-E) Allelic loss of Bif-1 suppresses lung perivascular tumor formation in Eμ-Myc–transgenic mice. The lungs (D) and livers (data not shown) from lymphomatous Eμ-Myc/Bif-1+/+ (n = 34; ii), Eμ-Myc/Bif-1+/− (n = 38; iii), Eμ-Myc/Bif-1−/− (n = 8; iv), and control Bif-1+/+ mice (i) were stained with H&E. The degree of lymphoblast infiltration to pulmonary alveolar septa (D) or hepatic sinusoids (E) and perivascular interstitial stroma expanded by lymphoblasts (D-E) were scored as follows: 0 = normal; 1 = < 5%; 2 = 6%-15%; 3 = 16%-40%; and 4 = > 40%. The lines represent median values. Statistical significance was determined using a 2-way ANOVA followed by a Scheffe posthoc test. *P < .05.
During the course of monitoring lymphoma development, we found that a significant portion of Eμ-Myc/Bif-1+/− mice displayed the signs of lymphoma, including hunched posture, ruffled fur, slow movement, and labored breathing, without developing palpable swollen lymph nodes. Pathologic analysis revealed that the thymi in these mice were abnormally enlarged despite the absence of massive enlargement of lymph nodes (Figure 1B). Statistical analyses revealed that the frequency of thymic lymphoma observed in Eμ-Myc/Bif-1+/− mice was significantly higher than in Eμ-Myc/Bif-1+/+ mice (33.3% vs 10.0%, respectively), suggesting that allelic loss of Bif-1 affects proper lymphoblast homing. Increased frequency of thymic lymphoma was also observed in Eμ-Myc/Bif-1−/− mice (25.0%). Differences in the frequency of thymic lymphoma between Eμ-Myc/Bif-1+/+ and Eμ-Myc/Bif-1−/− mice did not reach statistical significance; however, this may be attributable to the decreased survival and thus lower availability of Eμ-Myc/Bif-1−/− mice. The majority of all Eμ-Myc mice, regardless of Bif-1 status, had developed splenomegaly at the onset of disease (Figure 1C). In addition, histological analyses showed that there were no obvious morphological differences among the lymphomatous spleens (data not shown).
Infiltration of lymphoblasts to the lung and the liver is frequently observed in lymphomatous Eμ-Myc–transgenic mice.22 Consistently, thickened alveolar septa infiltrated with blast cells were observed in the lungs of Bif-1+/+ mice harboring the Eμ-Myc transgene (Figure 1D). Similar levels of blast cell infiltration were detected in the intravascular compartments of lungs isolated from lymphomatous Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− mice. However, significant inhibition of perivascular clustering of blast cells in the lung was observed in mice lacking 1 Bif-1 allele, suggesting that Bif-1 regulates lymphoblast infiltration to certain types of tissues. In contrast, tumor infiltration in both the intravascular and perivascular regions of the liver was not affected by hemizygous or homozygous deletion of Bif-1 (Figure 1E).
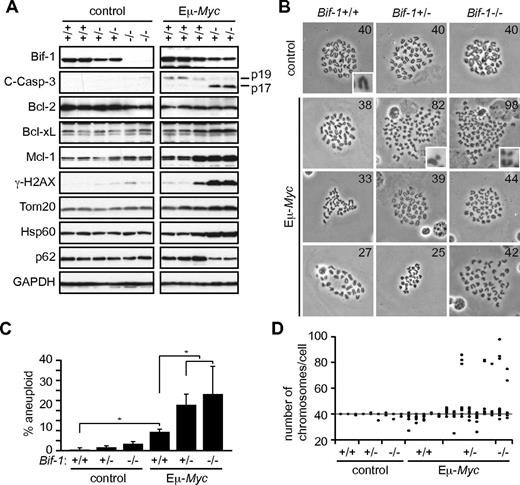
Bif-1 haploinsufficiency results in the up-regulation of Mcl-1 and suppresses caspase-3 activation in Eμ-Myc–induced lymphomatous lymph nodes
To determine whether the suppression of apoptosis is the mechanism by which Bif-1 deficiency accelerates Eμ-Myc-induced lymphomagenesis, axillary lymph nodes were collected after the onset of lymphoma and activation of caspase-3 was determined by monitoring caspase-3 processing. As shown in Figure 2A and B, hemizygous or homozygous deletion of Bif-1 significantly suppressed caspase-3 processing in lymphomatous lymph nodes. Similar results were obtained by immunohistochemical analyses (Figure 2C). In contrast, the expression of proliferating cell nuclear antigen was not affected by loss of Bif-1 (Figure 2D), suggesting that the suppression of apoptosis, rather than enhanced cell proliferation, is a mechanism through which the haplodeficiency of Bif-1 promotes Eμ-Myc–induced lymphomagenesis. In agreement with these data, we showed previously that stable knockdown of Bif-1 delays mitochondrial apoptosis and enhances the tumorigenicity of HeLa cells without affecting their proliferation rate.20
Bif-1 haploinsufficiency up-regulates Mcl-1 expression and suppresses caspase-3 activation in Eμ-Myc–induced tumors. Lymphomatous axillary lymph nodes were extirpated from each Eμ-Myc–transgenic mouse at the onset of disease. (A) Tissue lysates were prepared using a Tissuemiser homogenizer and analyzed by immunoblotting using the indicated antibodies. Representative blots of Eμ-Myc/Bif-1+/+ (n = 37), Eμ-Myc/Bif-1+/− (n = 38), and Eμ-Myc/Bif-1−/− (n = 8) tumors in addition to control Bif-1+/+ and Bif-1+/− lymph nodes are shown. (B) The intensity of cleaved-Caspase-3 (C-Casp-3, p17), Mcl-1, and p62 were quantified by densitometry using Quantity One software and are shown as a box plot. To compare the signal intensities between the different gels, the intensity of each blot was adjusted to the value of untreated wild-type mouse embryonic fibroblast lysates, which were loaded on every gel. Statistical significance was determined using 2-way ANOVA followed by the Scheffe posthoc test. *P < .05. (C-D) Tissues subjected to immunohistochemical analyses using the indicated antibodies. The number of proliferating cell nuclear antigen (PCNA)–positive cells per field (400×) was counted using a light microscope and is shown as a box plot in panel D. A total of 7451, 7251, and 9947 PCNA+ cells were counted from 3 Eμ-Myc/Bif-1+/+, 3 Eμ-Myc/Bif-1+/−, and 4 Eμ-Myc/Bif-1−/− mice, respectively.
Bif-1 haploinsufficiency up-regulates Mcl-1 expression and suppresses caspase-3 activation in Eμ-Myc–induced tumors. Lymphomatous axillary lymph nodes were extirpated from each Eμ-Myc–transgenic mouse at the onset of disease. (A) Tissue lysates were prepared using a Tissuemiser homogenizer and analyzed by immunoblotting using the indicated antibodies. Representative blots of Eμ-Myc/Bif-1+/+ (n = 37), Eμ-Myc/Bif-1+/− (n = 38), and Eμ-Myc/Bif-1−/− (n = 8) tumors in addition to control Bif-1+/+ and Bif-1+/− lymph nodes are shown. (B) The intensity of cleaved-Caspase-3 (C-Casp-3, p17), Mcl-1, and p62 were quantified by densitometry using Quantity One software and are shown as a box plot. To compare the signal intensities between the different gels, the intensity of each blot was adjusted to the value of untreated wild-type mouse embryonic fibroblast lysates, which were loaded on every gel. Statistical significance was determined using 2-way ANOVA followed by the Scheffe posthoc test. *P < .05. (C-D) Tissues subjected to immunohistochemical analyses using the indicated antibodies. The number of proliferating cell nuclear antigen (PCNA)–positive cells per field (400×) was counted using a light microscope and is shown as a box plot in panel D. A total of 7451, 7251, and 9947 PCNA+ cells were counted from 3 Eμ-Myc/Bif-1+/+, 3 Eμ-Myc/Bif-1+/−, and 4 Eμ-Myc/Bif-1−/− mice, respectively.
Inactivation of the ARF-MDM2-p53 pathway or dysregulation of antiapoptotic Bcl-2 family proteins are frequently observed in lymphomatous Eμ-Myc–transgenic mice23,24 and thus are believed to be major contributors for Myc-driven tumorigenesis. Therefore, we next examined the expression profiles of these apoptosis-related proteins in lymphomatous tumors. Consistent with previous studies,23,24 up-regulation of ARF and Bcl-xL was detected in Eμ-Myc/Bif-1+/+ tumors compared with normal lymph nodes obtained from control wild-type mice (Figure 2A). For all tumors tested, up-regulation of p53 was rarely observed. Although the Myc-induced suppression of Bcl-2 is reportedly disabled during lymphomagenesis,24 the expression levels of Bcl-2 in the lymphomatous lymph nodes remained low compared with the control wild-type mouse lymph nodes. Despite significant reduction of caspase-3 processing, the expression levels of these antiapoptotic proteins in Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− tumors were comparable to those in Eμ-Myc/Bif-1+/+ tumors (Figure 2A-B). Interestingly, however, loss of Bif-1 significantly enhanced the expression of Mcl-1, another member of the antiapoptotic Bcl-2 protein family. Furthermore, the Eμ-Myc–induced down-regulation of p62, an autophagic adaptor protein that mediates cell survival and cytoprotective stress-response pathways, was significantly suppressed by deletion of a Bif-1 gene, supporting previous reports that Bif-1 loss suppresses autophagy.21,25,26 Because overexpression of Myc has been shown to induce autophagy27 and p62 is an autophagic substrate, inhibition of an efficient autophagic degradation may be responsible for the sustained expression of p62 in lymphomatous tumors in Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− mice.
Bif-1 haploinsufficiency promotes chromosomal instability during Myc-induced neoplastic transformation
To determine whether Bif-1 haploinsufficiency suppresses premalignant tumor cell death under the selection pressure, prelymphomatous Eμ-Myc mouse spleens in which overproliferated, yet not malignant, polyclonal lymphoblasts are predominantly present,28 were extirpated before the onset of disease and subjected to immunoblot analyses. We detected a slight activation of caspase-3 in prelymphomatous Eμ-Myc/Bif-1+/+ cells compared with control splenocytes isolated from age-matched Bif-1+/+ mice (Figure 3A). Allelic loss of Bif-1 did not suppress, but rather enhanced, caspase-3 cleavage in prelymphomatous Eμ-Myc splenocytes despite the increased levels of antiapoptotic Bcl-xL and Mcl-1 proteins detected in these cells.
Bif-1 haploinsufficiency promotes Myc-induced genomic instability in prelymphomatous splenocytes. Spleens were isolated from 9-week-old prelymphomatous Eμ-Myc–transgenic and age-matched control mice with the indicated Bif-1 alleles. (A) Tissue lysates were subjected to immunoblot analyses using the indicated antibodies. (B-C) The karyotypes of Bif-1+/+ (n = 151 from 3 mice), Bif-1+/− (n = 153 from 3 mice), Bif-1−/− (n = 154 from 3 mice), Eμ-Myc/Bif-1+/+ (n = 268 from 5 mice), Eμ-Myc/Bif-1+/− (n = 384 from 7 mice), and Eμ-Myc/Bif-1−/− (n = 127 from 2 mice) splenocytes were analyzed. At least 50 metaphases were evaluated for each mouse. Representative images and magnified images of chromosomes are shown in panel B and the insets in panel B, respectively. The percentage of aneuploidy and the number of chromosomes per cell are shown in panels C (mean ± SD) and D, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .01.
Bif-1 haploinsufficiency promotes Myc-induced genomic instability in prelymphomatous splenocytes. Spleens were isolated from 9-week-old prelymphomatous Eμ-Myc–transgenic and age-matched control mice with the indicated Bif-1 alleles. (A) Tissue lysates were subjected to immunoblot analyses using the indicated antibodies. (B-C) The karyotypes of Bif-1+/+ (n = 151 from 3 mice), Bif-1+/− (n = 153 from 3 mice), Bif-1−/− (n = 154 from 3 mice), Eμ-Myc/Bif-1+/+ (n = 268 from 5 mice), Eμ-Myc/Bif-1+/− (n = 384 from 7 mice), and Eμ-Myc/Bif-1−/− (n = 127 from 2 mice) splenocytes were analyzed. At least 50 metaphases were evaluated for each mouse. Representative images and magnified images of chromosomes are shown in panel B and the insets in panel B, respectively. The percentage of aneuploidy and the number of chromosomes per cell are shown in panels C (mean ± SD) and D, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .01.
Inhibition of autophagy by allelic loss of Beclin 1 or homozygous deletion of Atg5 promotes metabolic stress–induced chromosomal instability and mammary tumorigenesis.29 Because Myc activation induces autophagy27 and allelic loss of Bif-1 significantly increased p62 expression (Figure 2B), the elevated expression of antiapo-ptotic proteins in prelymphomatous Eμ-Myc/Bif-1+/− cells may be caused indirectly by enhanced accumulation of genomic damage. To determine whether Bif-1 haploinsufficiency promotes the induction of DNA damage during Myc-driven lymphomagenesis, we first examined the accumulation of γ-H2AX, a well-established DNA double-strand break marker. A strong accumulation of γ-H2AX signals was detected in prelymphomatous Eμ-Myc/Bif-1+/− spleens (Figure 3A). Furthermore, a slight accumulation of γ-H2AX expression was detected in Bif-1−/− spleens in the absence of the Eμ-Myc transgene, suggesting that Bif-1 deficiency itself leads to the induction of DNA damage.
To determine whether the enhanced DNA double-strand breaks present in Bif-1–deficient cells leads to chromosomal aberrations, we next analyzed the karyotypes of splenocytes prepared from prelymphomatous Eμ-Myc mouse spleens. Consistent with the γ-H2AX data (Figure 3A), the frequency of aneuploidy induced by Myc was significantly increased by hemizygous and homozygous deletion of Bif-1 (Figure 3B-C). Although the aneuploidy observed in Eμ-Myc/Bif-1+/+ splenocytes was accompanied by the deletion of chromosomes, a large portion of Eμ-Myc/Bif-1+/− splenocytes (32 of 43 aneuploid cells) and Eμ-Myc/Bif-1−/− splenocytes (22 of 23 aneuploid cells) contained amplified numbers of chromosomes that were frequently accompanied by double-minute chromosomes (Figure 3B and D). No statistical significance in the rate of aneuploidy was observed between Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− mice. These results demonstrate that Bif-1 haploinsufficiency promotes chromosomal instability during Eμ-Myc–induced lymphomagenesis.
Bif-1 haploinsufficiency results in the accumulation of mitochondria in prelymphomatous Eμ-Myc cells
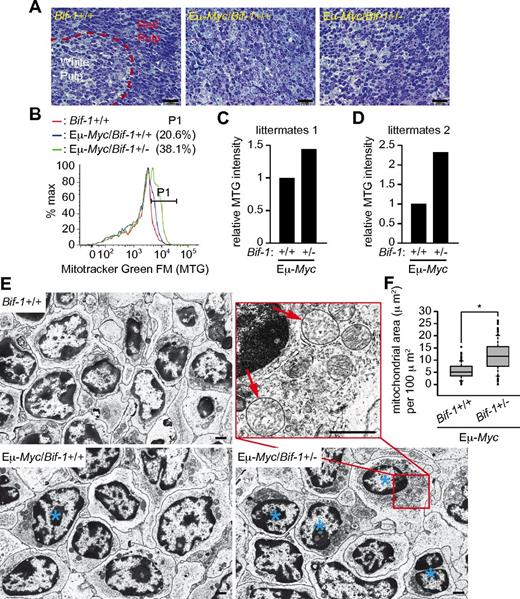
In addition to enhanced DNA damage induction, a negative correlation between expression of Bif-1 and accumulation of mitochondrial marker proteins (Tom20, a major component of the translocase of OMM complex and Hsp60, a mitochondrial matrix chaperonin) was also detected in the prelymphomatous Eμ-Myc transgenic spleen (Figure 3A). To determine whether Bif-1 haploinsufficiency promotes mitochondrial accumulation during Myc-induced neoplastic transformation, prelymphomatous splenocytes were isolated from littermates of Eμ-Myc/Bif-1+/− and Bif-1+/+ matings. Histological analyses revealed that the distinction between the red and white pulps was largely lost and monotonous plasmacytoid cells had accumulated in both Bif-1+/+ and Bif-1+/− spleens in the presence of an Eμ-Myc gene (Figure 4A), indicating that Bif-1 homozygosity is not required for Myc-induced overproliferation at the premalignant stage. The Myc-induced increase in intensity of the mitochondrial dye MitoTracker Green FM was further enhanced on hemizygous deletion of Bif-1 (Figure 4B-C). Similar results were obtained using another pair of littermates (Figure 4D). Furthermore, electron microscopic analyses showed that mitochondrial mass was indeed increased by Bif-1–hemizygous deletion (Figure 4E-F). Swollen mitochondria were occasionally observed in Eμ-Myc/Bif-1+/− cells (red arrows in Figure 4E). These results clearly indicate that Bif-1 is required for the maintenance of proper mitochondrial mass during Myc-induced lymphomatous transformation.
Bif-1 haploinsufficiency results in an increase in mitochondrial mass at premalignant stage of Myc-induced lymphoma. Prelymphomatous spleens were isolated from male littermate mice at the age of 12 weeks (panels A-C and E-F) and 10 weeks old (panel D). (A) Tissues were divided into 2 parts and a portion processed for electron microscopic analysis was stained with methylene blue and Azure II. (B) Splenocytes were prepared from another half portion of tissue, stained with MitoTracker Green FM for 30 minutes, and analyzed by flow cytometry. The percentages of cells with increased numbers of mitochondria by Myc are shown. (C-D) Geometric means of MitoTracker Green FM signals quantified and shown as relative intensities. (E) Tissue blocks in panel A were sectioned and analyzed by electron microscopy. The scale bars represent 1 μm. (F) Total mitochondrial area per cytoplasmic area in Eμ-Myc/Bif-1+/+ (n = 136) and Eμ-Myc/Bif-1+/− (n = 129) cells in panel E was quantified using ImageJ software and is shown as a box plot. The lines, boxes, and error bars represent median values, 25-75 percentiles, and 10-90 percentiles, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .0001.
Bif-1 haploinsufficiency results in an increase in mitochondrial mass at premalignant stage of Myc-induced lymphoma. Prelymphomatous spleens were isolated from male littermate mice at the age of 12 weeks (panels A-C and E-F) and 10 weeks old (panel D). (A) Tissues were divided into 2 parts and a portion processed for electron microscopic analysis was stained with methylene blue and Azure II. (B) Splenocytes were prepared from another half portion of tissue, stained with MitoTracker Green FM for 30 minutes, and analyzed by flow cytometry. The percentages of cells with increased numbers of mitochondria by Myc are shown. (C-D) Geometric means of MitoTracker Green FM signals quantified and shown as relative intensities. (E) Tissue blocks in panel A were sectioned and analyzed by electron microscopy. The scale bars represent 1 μm. (F) Total mitochondrial area per cytoplasmic area in Eμ-Myc/Bif-1+/+ (n = 136) and Eμ-Myc/Bif-1+/− (n = 129) cells in panel E was quantified using ImageJ software and is shown as a box plot. The lines, boxes, and error bars represent median values, 25-75 percentiles, and 10-90 percentiles, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .0001.
Loss of Bif-1 abrogates mitochondrial clearance by autophagy
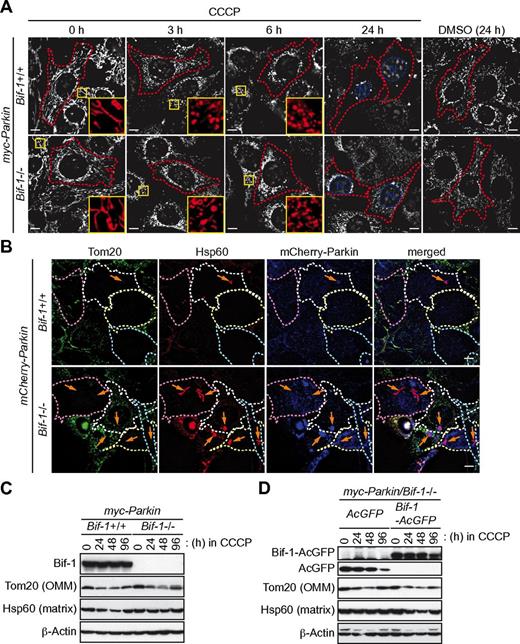
Autophagy is the central process for the maintenance of mitochondrial quality and quantity.30 To determine whether Bif-1 is responsible for the degradation of mitochondria, we took advantage of the Parkin-overexpression system, followed by treatment with a mitochondrial uncoupler, CCCP, a system that is widely used to study autophagy-dependent clearance of damaged mitochondria.31 After treatment with the mitochondrial uncoupler, the E3-ubiquitin ligase Parkin translocates from the cytosol to mitochondria, where it mediates the ubiquitination of a variety of OMM proteins to promote autophagic clearance of damaged mitochondria.30 After treatment with CCCP, mitochondria labeled with MitoTracker CMTMRos were fragmented and accumulated in the perinuclear region of myc-Parkin–expressing cells regardless of the expression of Bif-1 (Figure 5A). Although a significant portion of myc-Parkin/Bif-1+/+ cells partially or completely lost mitochondrial signals after 24 hours of treatment with CCCP, such a reduction of mitochondria was not observed in cells lacking Bif-1. Consistent with these observations, we found that both endogenous Tom20 and Hsp60 signals were fragmented and accumulated in the perinuclear region of both mCherry-Parkin/Bif-1+/+ and mCherry-Parkin/Bif-1−/− cells after 6 hours of treatment with CCCP (supplemental Figure 2). Moreover, both Tom20 and Hsp60 signals were partially or completely lost in Parkin-overexpressing wild-type cells after 24 hours of CCCP treatment (Figure 5B). We also found that Bif-1 loss abrogated CCCP-induced reduction of Hsp60, but not Tom20 signals. Similarly, Bif-1–dependent degradation of Hsp60, but not Tom 20, was detected in Parkin-overexpressing cells after treatment with CCCP, as shown by immunoblot analysis (Figure 5C). The decreased expression of mitochondrial proteins was restored 96 hours after the treatment, suggesting that mitochondria were regenerated. The inhibition of Parkin-dependent degradation of mitochondrial matrix markers in Bif-1–deficient cells was completely rescued by the restoration of Bif-1 expression (Figure 5D and supplemental Figure 3A). Because Parkin promotes the autophagy-independent degradation of OMM proteins through the ubiquitin-proteasome system after treatment with CCCP,32-34 these results clearly indicate that Bif-1 specifically regulates mitochondrial degradation through autophagy. Because Bif-1 is also known to accumulate in mitochondria on the induction of mitochondrial apoptosis20,35 and CCCP-induced mitochondrial degradation in mouse embryonic fibroblasts is dependent on Parkin32 (supplemental Figure 3B), Bif-1 may play a role in the mitochondrial translocation of Parkin on CCCP treatment. However, mitochondrial signals were extensively colocalized with mCherry-Parkin regardless of Bif-1 expression in response to CCCP (white arrows in supplemental Figure 2), thus suggesting that Bif-1 acts downstream of Parkin to regulate autophagy-dependent mitochondrial clearance (mitophagy).
Loss of Bif-1 suppresses mitophagy. Bif-1+/+ and Bif-1−/− mouse embryonic fibroblasts were transduced with lentiviruses encoding myc-Parkin or mCherry-Parkin and selected with puromycin for 5 days. (A) The resultant stable clones were prestained with MitoTracker CMTMRos, treated with 30μM CCCP or control DMSO for the indicated periods of time, and analyzed by fluorescent microscopy. Magnified images are shown in the insets. (B) Cells were treated with 30μM CCCP for 0, 6, and 24 hours and immunostained for Tom20 and Hsp60. Data shown are representative of the 24-hour time point; images at the 0- and 6-hour time points are shown in supplemental Figure 2. Arrows represent Hsp60+ and Tom20− structures. (C-D) Cells treated with 30μM CCCP for the indicated periods of time were subjected to immunoblot analysis using the indicated antibodies. The scale bars in panels A and B represent 10 μm.
Loss of Bif-1 suppresses mitophagy. Bif-1+/+ and Bif-1−/− mouse embryonic fibroblasts were transduced with lentiviruses encoding myc-Parkin or mCherry-Parkin and selected with puromycin for 5 days. (A) The resultant stable clones were prestained with MitoTracker CMTMRos, treated with 30μM CCCP or control DMSO for the indicated periods of time, and analyzed by fluorescent microscopy. Magnified images are shown in the insets. (B) Cells were treated with 30μM CCCP for 0, 6, and 24 hours and immunostained for Tom20 and Hsp60. Data shown are representative of the 24-hour time point; images at the 0- and 6-hour time points are shown in supplemental Figure 2. Arrows represent Hsp60+ and Tom20− structures. (C-D) Cells treated with 30μM CCCP for the indicated periods of time were subjected to immunoblot analysis using the indicated antibodies. The scale bars in panels A and B represent 10 μm.
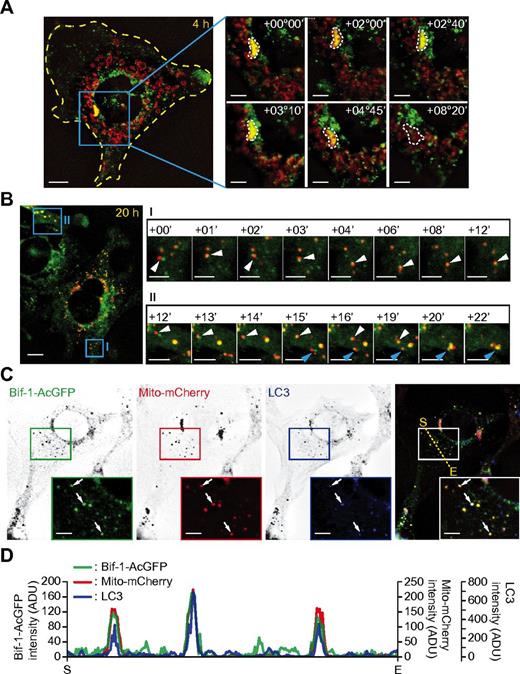
To gain further insight in the role of Bif-1 on mitophagy, myc-Parkin/Bif-1−/− cells transduced with Bif-1-AcGFP– and mito-mCherry–encoding lentiviruses were treated with CCCP and subjected to time-lapse fluorescence microscopic analyses. Consistent with our previous study,26 Bif-1 signals were detected diffusely in the cytoplasm in addition to the juxtanuclear region, but not at the mitochondria (data not shown). After treatment with CCCP, Bif-1 signals were time dependently accumulated on portions of fragmented and perinuclear-clustered mitochondria (Figure 6A). The signal intensities of Bif-1+ mitochondria were decreased over the incubation time period, suggesting that these structures were eventually degraded. Furthermore, short-interval imaging showed that Bif-1 strongly associated with fragmented mitochondria (Figure 6B). Some of the Bif-1+ mitochondria was colocalized with LC3 (Figure 6C-D), supporting the notion that Bif-1 regulates the autophagy-dependent clearance of mitochondria. To investigate this further, we next examined whether Bif-1 loss can also affect mitochondrial quantity control under ischemic condition, a physiologically relevant stress. Consistent with the data obtained from prelymphomatous Eμ-Myc splenocytes (Figure 3A), enhanced accumulation of mitochondrial markers and reduced degradation of p62 were observed in Bif-1−/− cells after exposure to hypoxic and low-nutrient conditions (supplemental Figure 4). Moreover, the induction of H2AX phosphorylation was enhanced by Bif-1 deficiency, also suggesting that Bif-1–mediated mitophagy may contribute to the suppression of DNA damage during metabolic stress.
CCCP induces Bif-1 and LC3 translocation to fragmented mitochondria.myc-Parkin/Bif-1−/− mouse embryonic fibroblasts were transduced with lentiviruses encoding Bif-1–AcGFP and Mito-mCherry. (A-B) The cells were treated with 30μM CCCP for 4 hours (A) or 20 hours (B) and then analyzed by time-lapse fluorescent microscopy at 5-minute (A) or 1-minute (B) intervals. The time stamps delineate the incubation time periods in hours (°) and minutes (′) after the initiation of time-lapse imaging. (C) Cells treated with 30μM CCCP for 20 hours, immunostained for LC3, and analyzed by fluorescence deconvolution microscopy. Magnified images are shown in the insets. Arrows indicate colocalization of Bif-1–AcGFP, Mito-mCherry, and LC3. (D) The fluorescence intensities along the dotted line in panel D were quantified using SlideBook Version 5.0 software. The values of the vertical axis represent fluorescence intensity units (ADU). The horizontal axis represents distance (S indicates start point; and E, end point). The scale bars represent 10 μm in the left panels in panels A and B and 5 μm in the right panels of time-lapse images in panels A through C.
CCCP induces Bif-1 and LC3 translocation to fragmented mitochondria.myc-Parkin/Bif-1−/− mouse embryonic fibroblasts were transduced with lentiviruses encoding Bif-1–AcGFP and Mito-mCherry. (A-B) The cells were treated with 30μM CCCP for 4 hours (A) or 20 hours (B) and then analyzed by time-lapse fluorescent microscopy at 5-minute (A) or 1-minute (B) intervals. The time stamps delineate the incubation time periods in hours (°) and minutes (′) after the initiation of time-lapse imaging. (C) Cells treated with 30μM CCCP for 20 hours, immunostained for LC3, and analyzed by fluorescence deconvolution microscopy. Magnified images are shown in the insets. Arrows indicate colocalization of Bif-1–AcGFP, Mito-mCherry, and LC3. (D) The fluorescence intensities along the dotted line in panel D were quantified using SlideBook Version 5.0 software. The values of the vertical axis represent fluorescence intensity units (ADU). The horizontal axis represents distance (S indicates start point; and E, end point). The scale bars represent 10 μm in the left panels in panels A and B and 5 μm in the right panels of time-lapse images in panels A through C.
Bif-1 regulates the maturation of nascent autophagosomes during mitophagy
Autophagy is initiated by the formation of isolation membranes (IMs)/phagophores that engulf cytoplasmic components and elongate, expand, and seal to form completed autophagosomes, which ultimately undergo maturation on fusion with endosomes and lysosomes for the degradation of the inner materials.36,37 To determine the step at which loss of Bif-1 suppresses mitophagy, Parkin-overexpressing cells were treated with CCCP for the indicated time periods and immunostained for Atg16L (IMs), LC3 (IMs and autophagosomes), and Lamp1 (lysosomes). As shown in supplemental Figure 5A and B, CCCP treatment induced the foci formation of Atg16L and LC3 in the close proximity of the clusters of Hsp60+ structures in wild-type cells. Lamp1 signals were also accumulated in close proximity to mitochondria, where they partially colocalized with LC3. Despite strong inhibition of mitochondrial loss, the CCCP-induced Atg16L and LC3 foci formation was not suppressed by knockout of Bif-1, indicating that Bif-1 is not required for the formation of nascent autophagosomes in response to CCCP treatment.
To further determine the effect of Bif-1 loss on mitophagy, the cells were subjected to electron microscopic analysis. We found that although a large portion of mitochondria were lost in CCCP-treated myc-Parkin/Bif-1+/+ cells, degradative autophagic vacuole (Avd) and nascent or immature autophagosome (Avi)–like structures were readily observable in the cytoplasm of these cells (Figure 7Ai). In stark contrast, many fragmented mitochondria were accumulated in the perinuclear region of CCCP-treated myc-Parkin/Bif-1−/− cells (Figure 7Aii). In agreement with a recent study,32 a large portion of these fragmented mitochondria were accompanied by either a partially or completely ruptured OMM structure (Figure 7Aii and iii and supplemental Figure 6B) that was distinct from the double-membrane structure of control DMSO-treated mitochondria (supplemental Figure 6A). Statistical analysis revealed that loss of Bif-1 significantly increased the number of total and ruptured mitochondria in the cytoplasm of CCCP-treated cells (Figure 7B), supporting our finding that Bif-1 is required for the autophagic clearance of damaged mitochondria. Consistent with the immunofluorescence data (supplemental Figure 5), the total number of CCCP-induced IM and Avi-like structures was not suppressed, but rather was slightly increased, by the loss of Bif-1 (Figure 7C). The IM/Avi-like structures (black arrows in Figure 7Aiii and iv and supplemental Figure 6B) in CCCP-treated myc-Parkin/Bif-1−/− cells were occasionally found to be associated with fragmented and/or ruptured mitochondria (double-arrowheads in Figure 7Aiii and iv and supplemental Figure 6B), although many of them remained associated with the endoplasmic reticulum (black arrowheads in Figure 7Aiii and iv and supplemental Figure 6B). Although numerous electron-dense lysosome-like structures were coaccumulated with ruptured mitochondria at the perinuclear region (Figure 7Aii and supplemental Figure 6B), the number of Avds was significantly reduced in Bif-1–deficient cells (Figure 7Av and D). These results suggest that Bif-1 regulates mitophagy by promoting the maturation of nascent autophagosomes.
Bif-1 plays a key role in the maturation of nascent autophagosomes during mitophagy. myc-Parkin–expressing mouse embryonic fibroblasts were treated with 30μM CCCP for 24 hours and subjected to electron microscopic analysis. (A) Representative images of Bif-1 wild-type and Bif-1–deficient cells are shown in subpanels i and ii. OMM-ruptured or OMM-preserved fragmented mitochondria that were closely associated with an isolation membrane are shown in subpanels iii and iv. An Avd-like structure observed in Bif-1–deficient cells is shown in subpanel v. Black arrowheads and arrows indicate endoplasmic reticulum (ER) membranes and ER-associated IMs, respectively. Open arrowheads, double-arrowheads, and arrows indicate Avd-like structures, fragmented mitochondria, and Avi-like structures, respectively. The scale bars represent 1 μm in subpanels i and ii and 0.5 μm in subpanels iii through v. The number of ruptured mitochondria and total mitochondria (OMM-ruptured and OMM-preserved; B), ER-associated IM/Avi-like structures and total IM/Avi-like structures (ER-associated and ER-nonassociated; C), and Avd-like structures (D) per cell were counted, normalized with cytoplasmic area (100 μm2) and are shown as box plots. The lines, boxes, and error bars represent median values, 25-75 percentiles, and 10-90 percentiles, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .0001; **P = .0227 in panel C and P = .0018 in panel D.
Bif-1 plays a key role in the maturation of nascent autophagosomes during mitophagy. myc-Parkin–expressing mouse embryonic fibroblasts were treated with 30μM CCCP for 24 hours and subjected to electron microscopic analysis. (A) Representative images of Bif-1 wild-type and Bif-1–deficient cells are shown in subpanels i and ii. OMM-ruptured or OMM-preserved fragmented mitochondria that were closely associated with an isolation membrane are shown in subpanels iii and iv. An Avd-like structure observed in Bif-1–deficient cells is shown in subpanel v. Black arrowheads and arrows indicate endoplasmic reticulum (ER) membranes and ER-associated IMs, respectively. Open arrowheads, double-arrowheads, and arrows indicate Avd-like structures, fragmented mitochondria, and Avi-like structures, respectively. The scale bars represent 1 μm in subpanels i and ii and 0.5 μm in subpanels iii through v. The number of ruptured mitochondria and total mitochondria (OMM-ruptured and OMM-preserved; B), ER-associated IM/Avi-like structures and total IM/Avi-like structures (ER-associated and ER-nonassociated; C), and Avd-like structures (D) per cell were counted, normalized with cytoplasmic area (100 μm2) and are shown as box plots. The lines, boxes, and error bars represent median values, 25-75 percentiles, and 10-90 percentiles, respectively. Statistical significance was determined using a Wilcoxon rank-sum test with continuity correction. *P < .0001; **P = .0227 in panel C and P = .0018 in panel D.
Discussion
In the present study, we provide the evidence that Bif-1 plays a key role in autophagy-dependent clearance of mitochondria and functions as a haploinsufficient tumor suppressor to prevent malignant transformation of Eμ-Myc transgenic pre-B cells. Our data show that Bif-1 haploinsufficiency results in the attenuation of caspase-3 activation in Eμ-Myc+ lymphoma cells (Figure 2A-C). In agreement with this finding, Bif-1 was originally discovered as a Bax activator to promote mitochondrial apoptosis in response to a variety of cytotoxic stresses.20,38 Bax loss has been shown to impair Myc-induced apoptosis and to accelerate Eμ-Myc–induced lymphomagenesis in mice.39 Therefore, one can speculate that suppression of the Bax-mediated apoptotic pathway could be the mechanism behind the inhibition of Eμ-Myc–induced caspase-3 activation by allelic loss of Bif-1. However, our data in prelymphomatous Eμ-Myc transgenic spleens revealed that Bif-1 haplodeficiency does not suppress, but rather promotes, caspase-3 activation (Figure 3A), suggesting that Bif-1 loss itself may not be sufficient to suppress the induction of apoptosis in overproliferated prelymphomatous cells under oncogenic/metabolic stresses. In contrast, we found that Bif-1 haploinsufficiency resulted in an increase in mitochondrial mass, the promotion of chromosomal instability, and the up-regulation of antiapoptotic proteins such as Mcl-1. Therefore, we propose that loss of Bif-1 enables tumor cells to adapt to selection pressures by enhancing chromosomal instability and up-regulating antiapoptotic Bcl-2 family proteins. Alternatively, Bif-1 haploinsufficiency may promote the survival of tumor cells through the up-regulation of proteins downstream of the mitochondrial pathway to inhibit the activity of cleaved mature caspases. Nonetheless, a significant reduction of cleaved caspase-3 accompanied by the up-regulation of Mcl-1 was observed in the majority of Eμ-Myc/Bif-1+/− and Eμ-Myc/Bif-1−/− cells after the onset of lymphoma, suggesting that attenuation of the mitochondrial pathway is likely a main cause of the acceleration of lymphomagenesis by Bif-1 haploinsufficiency.
Using Parkin-mediated mitophagy as a model, we also show in the present study that Bif-1 is required for autophagy-dependent clearance of damaged mitochondria. Because the accumulation of damaged mitochondria by impairment of autophagy results in the promotion of reactive oxygen species generation and DNA damage accumulation,12 failure of proper mitochondrial clearance is likely the mechanism behind the promotion of chromosomal instability by Bif-1 loss in Myc-induced lymphoma cells. However, we do not exclude the possibility that some other mechanisms may also contribute to Bif-1 haploinsufficiency-induced chromosomal instability. Interestingly, a recent study has shown that UVRAG, which bridges Bif-1 association with Beclin 1,21 regulates the activation of DNA-PK and promotes DNA double-strand-break repair in an autophagy-independent manner.40 Further studies are warranted to determine whether Bif-1 also regulates chromosomal instability and tumorigenesis through an UVRAG-dependent and autophagy-independent mechanism.
Although we and others have previously reported that Bif-1 forms a complex with Beclin 1 through UVRAG to regulate autophagosome formation induced by nutrient starvation or in models of Parkinson disease,21,25 the results of the present study showed that Bif-1 loss does not suppress, but rather accumulates, the IMs and the Avi-like structures during CCCP treatment. It has been reported that UVRAG promotes autophagy, not only by interacting with the Beclin 1-Vps34 complex at the autophagosome formation stage,41 but also by accelerating the maturation of endosomes and autophagosomes through the interaction with the c-Vps complex.42 Indeed, we found herein that the number of Avd-like structures induced by CCCP was slightly but significantly reduced by the loss of Bif-1 (Figure 7D). Many of the IMs/Avi-like structures observed in CCCP-treated Bif-1–deficient cells remained associated with the ER (Figure 7C). It has been shown previously that the IM, but not the completed autophagosome, frequently associates with the endoplasmic reticulum.43,44 Therefore, in this scenario, Bif-1 may play a key role in the completion of nascent autophagosome formation during mitophagy.
Our results also unveiled unexpected roles of Bif-1 in embryonic development and tumor cell homing in response to Myc dysregulation. It has been reported that a fraction of Eμ-Myc transgenic mice overexpressing Mcl-1 (VavP-Mcl-1/Eμ-Myc) undergo perinatal lethality.45 Moreover, in lymphomatous VavP-Mcl-1/Eμ-Myc mice, lymphoma cells were found to infiltrate and thicken the alveolar walls in the lungs, but not to accumulate in solid masses, a result similar to that observed by hemizygous deletion of Bif-1 in Eμ-Myc–transgenic mice. Therefore, up-regulation of Mcl-1 expression could be a mechanism behind the embryonic lethality and/or misregulated lymphoma cell homing by Bif-1 deficiency.
In summary, the results of the present study have important implications for the prevention and treatment of cancer because they offer novel insight into the role of mitophagy in cell survival, chromosomal integrity, and evasion of apoptosis during Myc-driven malignant transformation of hematopoietic cells. Our study provides an excellent model system for understanding the importance of mitochondrial quality control in embryonic development under oncogenic stress and tumor cell homing.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Arakawa, S. Shimizu, C. Kishi, N. Mizushima, and R. Meyers for technical advice or assistance on electron microscopy; E. L. Eskelinen for interpretation of the EM data; Q. Zhong for technical assistance on immunohistochemistry; and C. Meyerkord and K. Runkle for critical reading of the manuscript.
This work was supported by grants from National Institutes of Health (CA82197 and CA129682).
National Institutes of Health
Authorship
Contribution: Y.T. and T.H. performed most of the experiments and analyzed the data; T.K.C. conducted the histological and immunohistochemical analyses; J.L. performed the statistical analyses; N.D., J.M.S., and Y.I. assisted with the experiments; M.M.Y. assisted with the preparation of the manuscript; S.P. assisted with maintaining the mice; and Y.T. and H.-G.W. designed the experiments, prepared the figures, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hong-Gang Wang, Department of Pharmacology, The Pennsylvania State University, 500 University Dr, Hershey, PA 17033; e-mail: huw11@psu.edu.
References
Author notes
Y.T. and T.H. contributed equally to this work.