Key Points
NPM1 RT-PCR levels >1% are associated with poor overall and disease-free survival in AML patients treated with chemotherapy.
NPM1 MRD levels >10% are associated with poor overall and disease-free survival in AML patients after allogeneic transplantation.
Abstract
Mutations of the NPM1 gene (NPM1mut) are among the most common genetic alterations in acute myeloid leukemia and are suitable for minimal residual disease detection. We retrospectively investigated the prognostic impact of NPM1mut-based minimal residual disease detection from bone marrow for development of relapse by using a newly developed real-time polymerase chain reaction based on locked nucleic acid–containing primers in 174 patients, 155 of whom were treated within prospective protocols. The prognostic value of 5 cutoff values after completion of treatment or after allogeneic transplantation was studied by using cause-specific hazard models. Subsequent validation using cross-validated partial likelihood analysis revealed that an increase of more than 1% NPM1mut/ABL1 was most prognostic for relapse after chemotherapy, whereas an increase of more than 10% NPM1mut/ABL1 was most prognostic for relapse after allogeneic transplantation. Univariate and multivariate analysis of disease-free survival and overall survival revealed a significantly worse outcome in patients with >1% NPM1mut/ABL1 and >10% NPM1mut/ABL1, respectively, which remained significant after adjustment for FLT3–internal tandem duplication status. Our results in a large data set define and optimize cutoff values for early diagnosis of molecular relapse. These results may be especially important for defining triggers for early therapeutic intervention.
Introduction
In recent years, our understanding of the mechanisms responsible for the development and progression of disease in patients with acute myeloid leukemia (AML) has increased substantially. By characterizing AML on the basis of mutated genes, including Fms-like tyrosine kinase 3 (FLT3),1,2 nucleophosmin 1 (NPM1),3 and CCAAT enhancer-binding protein-α (CEBPA),4 important changes in prognostic disease classification have been made that affect AML patients with normal cytogenetics (CN-AML)—the largest subgroup (approximately 45%).5,6 Among these alterations, NPM1 mutations (NPM1mut) appear to be particularly important. Heterozygous mutations of NPM1 can be detected in approximately one-third of all AML patients and up to 60% of CN-AML patients.3,7 Thus, NPM1 gene mutations represent the most common molecular alteration found in adult AML to date. The NPM1 gene maps to chromosome 5q35 and contains 12 exons.8,9 It is chromosomally translocated in certain tumor types, resulting in the formation of fusion proteins that retain the N-terminus of NPM1.10 Heterozygous mutations affecting exon 12 involving the C-terminus are specific for AML.3 To date, more than 50 different mutations located in exon 12 of the NPM1 gene have been identified,11,12 however, three mutation types (A, B, and D) predominate and can be detected in about 95% of patients. Irrespective of the mutation type, all mutations cause a distinct sequence change at the C-terminus of NPM1, elongating the protein and substituting one or two tryptophan residues which, in turn, lead to the abnormal accumulation of the protein in the cytoplasm of NPM1-mutant cells.13 As a consequence of this altered localization, the NPM1 protein loses its important function in the regulation of several key proteins, including p14ARF.14 Both epidemiological and biological evidence suggest that NPM1 mutations are early causative alterations in leukemic stem and progenitor cells in the majority of patients with AML. This has led to the inclusion of NPM1 mutations together with CEBPA mutations as provisional entities in the most recent version of the World Health Organization classification.15 Various independent studies have shown a favorable impact of NPM1 gene mutations on clinical outcome in the CN-AML setting and a lack of FLT3–internal tandem duplication (FLT3-ITD) mutations.3,16-18 Nevertheless, a substantial proportion of patients with NPM1 mutations still show disease recurrence. In the relapse situation, several studies have shown stability of NPM1 gene alterations, implicating them as a potentially useful marker for minimal residual disease (MRD) detection.19,20 A number of sensitive methods for detecting NPM1 mutations have been reported.18,21 Real-time quantitative polymerase chain reaction (RT-qPCR)–based procedures appear to be useful because they can provide accurate information on the level of residual disease.21 By using qPCR methods, several groups have shown that relapse can be detected with considerable sensitivity.21-25 However, the number of reports in the context of controlled clinical studies is still limited, and important questions remain: What is the optimal cutoff for the diagnosis of molecular relapse? And what is the impact of NPM1 MRD for long-term prognosis? To this end, we developed an optimized qPCR procedure to detect the three most common mutations (types A, B, and D3 ) and analyzed the impact of residual mutant NPM1 in a large cohort of patients treated in two prospective clinical studies. Our results indicate that measurement of MRD as defined by mutant NPM1 can identify a group of patients with increased risk of recurrence and allows detection of relapse with high sensitivity in individual patients.
Patients and methods
Patients
We retrospectively investigated MRD in a total of 184 NPM1-mutant AML patients belonging to two patient cohorts. Patients with refractory disease (n = 10) were excluded. From the remaining 174 patients, a core population of 155 patients was enrolled onto two clinical trials of the Study Alliance Leukemia (SAL) (for further information on protocols and treatment regimens, see the supplemental Data). Of these 155 patients, 40 patients received allogeneic stem cell transplantation (allo-SCT) (Table 1). For the analysis of the relevance of NPM1 MRD after allo-SCT, we collected data for an additional set of 19 patients who received transplants and who had primary refractory disease or relapse but were not part of the above-mentioned clinical studies. Inclusion criterion for this analysis was the presence of an NPM1 mutation (type A, B, or D) at diagnosis. All patients gave written informed consent in accordance with the Declaration of Helsinki to participate in the study. Institutional review board approval was obtained for the study protocols and the additional molecular studies from the Dresden University of Technology.
Time points of MRD sample collection
Altogether, 1750 samples (bone marrow [BM], n = 933; peripheral blood [PB], n = 817) were analyzed and included in the statistical analysis; of these, 138 samples were paired BM and PB samples collected from individual patients at the same time point. By using data from this complete data set, we performed 4 separate subsample analyses—(1) receiver operating characteristic (ROC) analysis, (2) prognostic value of MRD after completion of treatment (excluding allo-SCT), (3) prognostic value of MRD after allo-SCT, and (4) prognostic value of MRD level at the time of the first complete response (CR1)—as detailed in the supplemental Data.
Molecular analysis
Initial screening for NPM1 mutations was performed as described previously.18 All samples that were positive for mutant NPM1 were then sequenced by using a locked nucleic acid (LNA) clamping strategy.26 For MRD quantification, an improved qPCR assay for the detection of the three most common NPM1 mutations (types A, B, and D) was developed. By using published assays,22 a high rate of unspecific amplification was observed for the most common variant (mutation type A; duplication of the bases TCTG). To overcome this unspecific amplification, LNA bases were incorporated into the primer sequences of the mutant-specific primer as illustrated in supplemental Figure A. A common generic probe and reverse primer were used for all reactions. PCR was performed on a LightCycler480 instrument (Roche, Mannheim, Germany) using 5′-nuclease chemistry. In the final optimized procedure, the 20-µL reaction mixture consisted of 4 µL of LightCycler TaqMan Master kit (Roche), 0.25 µM NPM1 mutation–specific primer, 0.3 µM common NPMs primer, 0.2 µM generic NPM1 probe, 12.5 µL of water, and up to 2 µL of complementary DNA (cDNA), prepared from 1 to 5 µg of total RNA as described.27 After an initial step of 95°C for 10 minutes, 50 cycles of a two-step PCR were performed, with 20 seconds at 94°C for denaturation and 1 minute at 66°C for annealing and elongation. Absolute quantification was performed based on standard curves by using cloned cDNAs of the individual mutants. Results were adjusted to ABL1 as the reference gene and expressed as percent NPM1mut/ABL1. A minimum ABL1 copy number of 1000 copies was required for inclusion of a sample; the median ABL1 copy number of all samples investigated was 30 000. All samples without a signal after 50 cycles were considered negative for NPM1mut. A reaction containing MV4-11 (wild-type NPM1 [wt-NPM1]) cells was run with every qPCR to control for unspecific amplification, along with appropriate positive and no-template controls.
Statistical analysis
The initial exploratory analysis to identify informative NPM1 cutoff points was performed with ROC curves using all data. However, this approach was problematic, since multiple observations per patient were treated as independent observations. To overcome this limitation, these findings were further explored by using Cox regression, and the most predictive model was identified by using the cross-validated partial likelihood (CVPL) procedure of Verweij and van Houwelingen.28 A more detailed description of the statistical methods and the underlying assumptions is given in the supplemental Data.
To visualize and estimate the median time for an event after exceeding the defined thresholds, Kaplan-Meier and cumulative incidence of relapse (CIR) curves were generated. Thus, the displayed numbers at risk in each Kaplan-Meier and CIR plot do not display the number of patients but the number of observations belonging to each cutoff category (exemplified in supplemental Figure 2). CIR was calculated according to Kalbfleisch and Prentice.29 Death and relapse were considered as competing events. CR and relapse were defined according to criteria reported by Cheson et al.29,31 Disease-free survival (DFS) was defined only for patients who achieved CR and was measured from the documented date of CR until date of relapse or last follow-up date available or death regardless of cause. Overall survival (OS) was measured from date of diagnosis until date of death, regardless of cause of death or date of last follow-up. Kaplan-Meier curves were calculated for DFS and OS. To adjust for potential confounding covariates, univariate and multivariate Cox proportional hazard models adjusted for the following variables were used: lactate dehydrogenase, white blood cell counts, FLT3-ITD status, and age. A two-sided P < .05 was regarded as significant. The distribution of categorical parameters was analyzed with the χ2 test.
Results
Sensitivity and specificity of RT-qPCR
Using the optimized RT-PCR conditions, one NPM1mut OCI-AML3 cell was detected in a background of 100 000 MV4-11 cells. For the other mutants for which no positive cells lines were available, we could demonstrate that a 5-fold log10 dilution of a mutant sample obtained at diagnosis in a background of wt-cDNA was detectable. Using the final conditions, none of the analyzed NPM1-wt materials (including cell lines as well as BM samples from lymphoma patients and healthy BM donors) tested positive.
Determining a prognostic threshold of NMP1mut/ABL 1% for relapse
The main objective of our analysis was to identify an NMP1mut/ABL1 percent level that would be the most informative for the occurrence of relapse after completion of treatment with regard to outpatients. Considering the kinetics of NPM1mut burden after achieving CR, ROC curves of 282 BM samples from 101 patients were used to assess the optimal cutoff value that yielded the best separation of patients into two groups in terms of relapse through maximizing Youden’s index. For the calculation of the index for a given patient, all samples within our predefined time intervals (30, 60, 90, 120, 180, and 360 days) were included in the analysis. Based on the maximization of Youden’s index, the threshold value of 1% NPM1mut/ABL1 represented the optimal sensitivity and specificity with the corresponding Youden’s index of 0.88 for the time point 90 days. Within 90 days, 10 patients relapsed, 9 of whom had an increase of more than 1%; thus, the sensitivity was 90%. According to this analysis, 23 measurements from 17 patients were denoted false positive, since these patients did not relapse until 90 days after observation. However, altogether, 27 patients in this cohort relapsed at 360 days. Thus, of the 17 false-positive patients, 9 relapsed after the observation time of 90 days, whereas 8 did not relapse (4 received an allogeneic transplant, 1 was given an autologous transplant, and 1 received an additional consolidation cycle). Thus, only two patients did not show relapse, although they had an increase of more than 1% and did not receive further documented treatment, indicating a much higher practical specificity. However, this also indicates the limitation of the ROC analysis in this situation, because it is restricted to certain time periods, and multiple samples of patients were used for this analysis. To overcome these limitations, we performed additional analyses based on five distinct cutoffs (0.01%, 1%, 2%, 5%, and 10% NMP1mut/ABL1). The choice of cutoffs was based on the findings of the ROC analysis (1%) and recently published data.24,25 This analysis was carried out by fitting a Cox regression model to our NPM1mut-level data, yielding a cause-specific hazard ratio (CHR) estimate for the MRD cutoffs. The analysis considered MRD after completion of treatment to identify patients at risk for relapse. We incorporated 208 BM samples from 60 patients (of whom 27 had relapsed) into this analysis. In doing so, the following risk groups were generated: NPM1mut/ABL1 >0.01% vs ≤0.01%; NPM1mut/ABL1 >1% vs ≤1%; NPM1mut/ABL1 >2% vs ≤2%; NPM1mut/ABL1 >5% vs ≤5%; and NPM1mut/ABL1 >10% vs ≤10% (Table 2 and supplemental Table 3). NPM1mut/ABL1 ≤0.01% included patients with no detectable MRD as well.
By maximizing CVPL, a level of 1% NPM1mut/ABL1 emerged as having the strongest indicative value for relapse (Table 2 and supplemental Table 4). Further analyses focusing on the risk groups NPM1mut/ABL1 >1% vs ≤1% were conducted. In addition to the MRD level (>1% vs ≤1%), we considered the remission status and checked whether the NPM1mut/ABL1 percentage at time of CR had an impact on the adjusted CHR (Table 2). We could not demonstrate that the MRD status at the time of CR1 (MRD ≤1% or ≤0.01%) was associated with the risk for relapse after adjustment for MRD information derived from later time points (Table 2). The CIR analyzed according to the Fine and Gray model for competing risks revealed that the probability for relapse increases with increasing MRD burden (Figure 1). The median time to relapse was 121 days (range, 70 to 172 days) for the MRD >1% NPM1mut/ABL1 subgroup and 66 days (range, 34 to 98 days) for patients exceeding the 10% NPM1mut/ABL1 cutoff after therapy (supplemental Table 4). The threshold of 1% could be confirmed in an analogous analysis when we put both BM and PB sources together (data not shown). A separate analysis for PB in this setting could not be initiated because of the low number of samples. Patients with MRD >1% NPM1/ABL1 levels also also showed inferior OS and DFS (Figure 3). Furthermore, a sensitivity analysis stratifying for the two SAL trials did not reveal different results for either the identified CHR or for DFS and OS (data not shown). To estimate the number of NPM1-mutant cells present in patients with a 1% NPM1mut/ABL1 level, we performed ultradeep next-generation amplicon sequencing on an IonTorrent PGM platform in 10 patients; qPCR results were between 0.8% and 2% NPM1mut/ABL1 using genomic DNA as source material. With a median of 160 000 reads performed per sample, the median NPM1-mutant:NPM1-wt allelic ratio detected was 0.016%, indicating that approximately 1 in 30 000 cells carried the mutation.
CIR for different NPM1mut/ABL1 cutoffs after completion of treatment with conventional chemotherapy and autologous SCT through follow-up. CIR analyzed by Gray’s regression model was affected by increasing MRD burden. CIR was calculated for the following NPM1mut/ABL1 cutoffs: (A) 0.01%, (B) 1%, (C) 1% and FLT3-ITD status, (D) 2%, (E) 5%, and (F) 10%.
CIR for different NPM1mut/ABL1 cutoffs after completion of treatment with conventional chemotherapy and autologous SCT through follow-up. CIR analyzed by Gray’s regression model was affected by increasing MRD burden. CIR was calculated for the following NPM1mut/ABL1 cutoffs: (A) 0.01%, (B) 1%, (C) 1% and FLT3-ITD status, (D) 2%, (E) 5%, and (F) 10%.
CIR for different NPM1mut/ABL1 cutoffs after allogeneic SCT beginning from the first MRD assessment post-SCT. CIR analyzed by Gray’s regression model indicates that the probability of relapse correlates with MRD burden. CIR was investigated for the following NPM1mut/ABL1 cutoffs: (A) 0.01%, (B) 1%, (C) 2%, and (D) 10%.
CIR for different NPM1mut/ABL1 cutoffs after allogeneic SCT beginning from the first MRD assessment post-SCT. CIR analyzed by Gray’s regression model indicates that the probability of relapse correlates with MRD burden. CIR was investigated for the following NPM1mut/ABL1 cutoffs: (A) 0.01%, (B) 1%, (C) 2%, and (D) 10%.
OS and DFS by 1% NPM1mut/ABL1 cutoff. MRD clearance or MRD reduction ≤1% after completion of study treatment translated clinically into a better (A) DFS and (B) OS. The subgroup with >1% MRD displayed a significantly worse outcome in comparison with the negative and <1% subgroups. Note that the DFS analysis started from the time of completion of the study treatment.
OS and DFS by 1% NPM1mut/ABL1 cutoff. MRD clearance or MRD reduction ≤1% after completion of study treatment translated clinically into a better (A) DFS and (B) OS. The subgroup with >1% MRD displayed a significantly worse outcome in comparison with the negative and <1% subgroups. Note that the DFS analysis started from the time of completion of the study treatment.
Impact of FLT3-ITD status on outcome
We then analyzed the influence of FLT3-ITD mutation status alone and the interaction with NPM1mut/ABL1 level by modeling the CHR using NPM1mut/ABL1 with a cutoff at 1% and FLT3-ITD and incorporating the interaction term of both factors (Table 2). FLT3-ITD by itself had a significant impact on relapse but the interaction of FLT3-ITD with the MRD level did not. The CIR for patients carrying the FLT3-ITD mutation and MRD >1% was increased in comparison with that of the other subgroups (Figure 1). The median time to relapse was 85 days.
NPM1mut/ABL1 >10% after SCT is associated with an increased rate of relapse
The relevance of increasing NPM1 MRD after SCT was investigated in a subset of patients (n = 59) after they received an allogeneic graft, among whom 11 patients experienced relapse, 8 died of causes other than relapse, and 40 remained in remission. In this situation, we again used all five cutoffs (0.01%, 1%, 2%, 5%, and 10%) previously explored after chemotherapy. For this cohort, samples from BM and PB (depending on their availability) were considered because of the low number of BM samples and the fact that when we put both sources together in the chemotherapy-treated group, the identified cutoff from BM was confirmed. The analysis was again carried out by estimation of the NPM1mut/ABL1-specific HR with Cox regression models. An increasing MRD burden exceeding the threshold of 10% NPM1mut/ABL1 displayed the strongest HR for relapse by maximizing CVPL (Table 2 and supplemental Table 4). The CIR analyzed according to the Fine and Gray model for competing risks indicated that patients with increasing NPM1mut/ABL1 are at higher risk of experiencing relapse (Figure 2). Prediction of outcome based on DFS and OS analysis demonstrated a significantly worse outcome for the MRD >10% NPM1mut/ABL1 subgroup (Figure 4). The median time to relapse for the MRD 0.01% NPM1mut/ABL1 and >1% NPM1mut/ABL1 subgroups was not reached. For the 10% NPM1mut/ABL1 subgroup, the median time to relapse was 68 days (Table 2).
DFS and OS according to MRD level after allogeneic SCT. After SCT both MRD >1% and >10% translated into a poor outcome with a significantly higher risk to develop relapse or die. (A-B) DFS after SCT analyzed for increasing NPM1mut/ABL1 >1% (A) and NPM1mut/ABL1 >10% (B). (C-D) OS after SCT for the corresponding subgroups.
DFS and OS according to MRD level after allogeneic SCT. After SCT both MRD >1% and >10% translated into a poor outcome with a significantly higher risk to develop relapse or die. (A-B) DFS after SCT analyzed for increasing NPM1mut/ABL1 >1% (A) and NPM1mut/ABL1 >10% (B). (C-D) OS after SCT for the corresponding subgroups.
Prognostic impact of depth of molecular remission in CR1
The impact of the level of residual molecular disease as measured by the NPM1mut/ABL1 ratio at the time of achieving CR1 on OS and DFS was assessed. As shown in Figure 5 and Table 3, patients with detectable MRD exceeding the >1% threshold showed a significantly worse DFS and OS in comparison with patients without detectable MRD. In a multivariate analysis adjusted for FLT3-ITD mutation status, lactate dehydrogenase, white blood cell count, and age at diagnosis, only the factor NPM1mut/ABL1 >1% at the time of CR1 remained statistically significant for DFS and OS (DFS: HR, 9.58; 95% CI, 2.04 to 45.01; P = .004. OS: HR, 5.81; 95% CI, 1.16 to 29.2; P = .032). The number of FLT3-ITD–positive patients in the cohorts NPM1neg, NPM1mut/ABL1 <1%, and NPM1mut/ABL1 >1% were 30%, 23%, and 48%, respectively (P = .10).
DFS and OS by MRD level at CR1. MRD clearance or MRD reduction ≤1% after achievement of CR after induction treatment was associated with improved (A) DFS and (B) OS. The subgroup with >1% MRD displayed a significantly worse outcome in comparison with the MRD-negative and <1% subgroups.
DFS and OS by MRD level at CR1. MRD clearance or MRD reduction ≤1% after achievement of CR after induction treatment was associated with improved (A) DFS and (B) OS. The subgroup with >1% MRD displayed a significantly worse outcome in comparison with the MRD-negative and <1% subgroups.
Comparison of different sample sources: BM vs PB
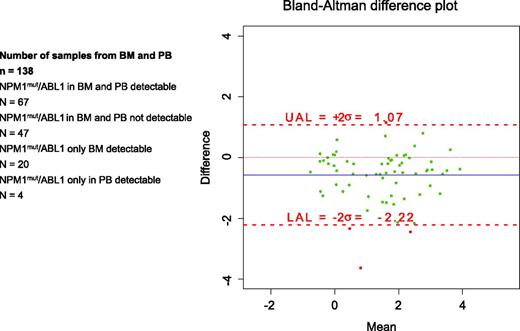
To investigate whether NPM1mut can be effectively monitored in PB, we compared MRD measurements in 138 pairs of BM and PB samples collected in parallel during the follow-up (Figure 6). MRD was detected in both BM and PB in 67 paired samples and in BM but not in PB in 20. It was undetectable in 47 sample pairs, and in the remaining 4 matched materials, it was detectable only in PB whereas BM was not. Since neither the correlation coefficient nor regression analysis is appropriate for assessing agreement between the two sources, we used the Bland-Altman difference plot.32 The differences between the two sources were plotted against the averages. The limits of agreement were 1.07% to −2.22% (Figure 6). The presence of MRD in PB was generally associated with higher levels of MRD in the corresponding BM sample; the median absolute difference between BM and PB was 0.8% (95% CI, 0.65% to 0.95%). Thus, in general, the median NPM1mut levels in PB samples were almost 1-log10 lower.
Assessing the limits of agreement between BM and PB from 138 samples by Bland-Altman difference plot. The differences between the two sources were plotted against their averages. The analysis revealed that assessment from PB may be 1.07% above or 2.22% below BM. UAL, upper analysis limit; LAL, lower analysis limit.
Assessing the limits of agreement between BM and PB from 138 samples by Bland-Altman difference plot. The differences between the two sources were plotted against their averages. The analysis revealed that assessment from PB may be 1.07% above or 2.22% below BM. UAL, upper analysis limit; LAL, lower analysis limit.
Discussion
RT-PCR for NPM1mut is an attractive tool for assessment of MRD because of the high prevalence of this alteration and its stability in relapse. As a consequence, several groups have established protocols for detecting this change. When we started to study this parameter, we tried to use the procedure first published by Gorello et al.21 Since NPM1mut is highly expressed on the messenger RNA (mRNA) level, we aimed to use RNA as source material. However, we observed a considerable level of unspecific amplification, which induced us to design an optimized assay, especially for the most common variant (type A; duplication of the bases TCTG). By inclusion of an LNA base in the mutant-specific primer, we could significantly improve the specificity of the reaction. Because of their altered ribose structure, LNA-containing oligonucleotides display remarkably increased thermodynamic stability and enhanced specificity. On the basis of these modified conditions, we achieved a routine level of sensitivity of one mutant cell in a background of 100 000 wt cells.
We then used this procedure to retrospectively investigate whether the detection of relapse is possible on the basis of increasing NPM1mut MRD after exceeding a certain cutoff. Our results confirm that NPM1mut-based MRD monitoring allows for identification of patients at an increased risk of relapse. The data are in good concordance with those from previous reports investigating comparable data sets, namely the data reported by Schnittger et al25 and Krönke et al,24 who emphasized that investigation of MRD is a valuable tool for the prediction of outcome in NPM1mut AML. Schnittger et al discriminated for a cutoff value of 0.01% NPM1mut/ABL1 at different time points during first-line treatment of two distinct subgroups differing in event-free survival (EFS) but not in the risk of relapse or OS. After SCT and during second-line therapy, different prognostic subgroups for EFS were defined by a cutoff value of 0.1% NPM1mut/ABL1. A major issue in this context is that EFS includes patients with primary refractory disease and patients who never achieved CR. We could not confirm the threshold of 0.01% NPM1mut/ABL1 for the patient cohort with first-line therapy. These discrepancies might be related to the different sampling intervals. Schnittger et al25 grouped the cohort into time intervals for MRD assessment (days 18 to 60, 61 to 120, 121 to 365, and after 1 year). This strategy is controversial because time points after initial diagnosis are not strictly connected to the stage of treatment. Our analysis rigorously focused on the development of MRD level after completion of conventional treatment to identify outpatients at an increased risk for relapse. Furthermore, affirming the value of 1% (corresponding to 100 copies NPM1mut/10 000 copies of ABL1) very closely matches the value reported by Krönke et al24 who used a different, arbitrary approach for the definition of a value of 200 copies NPM1mut/10 000 copies ABL1 (equivalent to 2% NPM1mut/ABL1) to be predictive for development of relapse. These authors addressed the question of whether quantitative NPM1mut reduction at time of CR1 affects DFS and OS. The aim of our study was to further extend this analysis and to provide a prognostic cutoff value for individual patient-specific outcome assessment after completion of treatment. By using two different statistical methods for patients undergoing chemotherapy, the same value of >1% NPM1mut/ABL1 could be confirmed. Because technical differences and individual methodologic aspects can make it difficult to compare different laboratories and different methods (eg, RT-PCR and flurorescent-activated cell sorting), we tried to determine the corresponding MRD cutoff on the cellular level. By using next-generation sequencing, we obtained a value of 0.016% NPM1mut alleles per NPMwt alleles, indicating that approximately 1 in 30 000 cells carried the mutation. Interestingly, this value corresponds well with the data obtained in acute lymphoblastic leukemia, in which a cutoff of 1 in 10 000 cells (10−4) is used to stratify patients at high and low risk after chemotherapy;33 this value is also the same order of magnitude as that recently reported for MRD based on flow cytometry in AML.34
After SCT, the best prognostic cutoff in our analysis was 10% NPM1mut/ABL1. Interestingly, although the data do not match on an absolute scale, Schnittger et al also reported a 10-fold higher cutoff value for relapse and transplanted patients in their analysis.25 In our study, we cannot rule out that the patients in our analysis were given immunotherapy such as cessation of the immunosuppressive regimen because of increasing MRD burden. Nevertheless, a graft-versus-leukemia reaction induced by immunogenicity of the mutant NPM1 protein might be an explanation for this observation, so that at lower MRD levels, the allogeneic graft is able to repress leukemia, but after crossing a certain MRD burden, the graft-versus-leukemia reaction is no longer sufficient for preventing relapse. It has been shown that mutant NPM1 can be recognized by human leukocyte antigen A1–restricted cytotoxic T lymphocytes; thus, it might be speculated that dislocation of the mutant NPM1 protein to the cytoplasm may promote the processing by the class I degradation pathway, thereby leading to human leukocyte antigen presentation and immune response.36-38
Replacement of BM sampling by PB has been a topic of debate in MRD studies, because the ease of sampling would enable a much more frequent sampling. We investigated the level of concordance for both sources. Although the level of MRD in PB is almost 1-log10 lower than that in BM, which is in line with previous reports,25 NPM1 mutations were detectable in the majority of samples. Thus, for clinical purposes in patients who are negative in BM, PB sampling at regular, probably shorter intervals than for BM might be appropriate. However, since the 1% cutoff value was established by using BM at certain decisive time points (eg, after completion of induction or after consolidation), BM sampling appears mandatory.
Our study indicates that in chemotherapy-treated patients, rising MRD levels >1% NPM1mut/ABL1 are associated with an increased risk for relapse. This of course raises the question of suitable treatment options. In a first observational series of 10 patients who were treated for increasing NPM1 MRD levels by using 5-azacytidine, we found good results35 which spurred the initiation of a prospective, interventional phase II study (NCT01462578). In addition, since mutant NPM1 seems to induce an antileukemic T-cell response, it could serve as a target in the setting of immunotherapy. Future strategies might therefore additionally use adoptive immunotherapy in case of reoccurrence of mutated NPM1 after SCT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the important contribution of all participating centers and physicians of the Study Alliance Leukemia (SAL) who assigned patients to the prospective SAL treatment protocols and provided samples. The authors also thank Marika Karger, Annica Schulze, and Marita Hartwig for excellent technical assistance.
This work was supported in part by grants BMBF 03WKBH2H and 01GU1108A (C.T.)
Authorship
Contribution: N.S. and C.T. designed the study, performed research, and wrote the manuscript; M.K. and J.S. performed statistical analysis; M.B., M.S., U.P., C.R., and G.E. wrote the clinical study protocols and treated patients; C.H. performed next-generation sequencing; O.L. designed the qPCR assay; and all authors read, edited, and approved the final manuscript. A complete list of the members of the investigators of the AML 2003 and AML 60+ trials appears in “Appendix.”
Conflict-of-interest disclosure: O.L. owns a company producing oligonucleotides and molecular diagnostic kits (TIB MOLBIOL GmbH, Berlin, Germany). C.T. co-owns a company that performs molecular diagnostics (AgenDix GmbH, Dresden, Germany), C.H. is employed by a company that performs molecular diagnostics (AgenDix GmbH, Dresden, Germany). The remaining authors declare no competing financial interests.
Correspondence: Christian Thiede, Medizinische Klinik und Poliklinik I, Universitätsklinikum Carl Gustav Carus der Technischen Universität, Fetscherstrasse 74, 01307 Dresden, Germany; e-mail: christian.thiede@uniklinikum-dresden.de.
Appendix
A. Schulz-Abelius, K. Friedrichsen (Klinikum Altenburg, Germany); R. Repp, G. Helm (Klinikum Bamberg, Germany); A. Kiani, A. Krost (Klinikum Bayreuth, Germany); E. Thiel, C.D. Baldus (Charité Campus Benjamin Franklin, Berlin, Germany); K. Possinger, D. Kühnhardt (Charité Campus Mitte Berlin, Germany); M. Görner, S. Probst (Städtische Kliniken, Klinikum Mitte Bielefeld, Germany); F. Weissinger, U. Krümpelmann (Krankenanstalten Gilead, Bielefeld, Germany); K.-H. Pflüger, C. Diekmann (Evangelische Diakonissenanstalt Bremen, Germany); J. Mayer, M. Protivankova (University Hospital Brno, Czech Republic); M. Hänel, R. Herbst (Klinikum Chemnitz, Germany); H.-J. Pielken, H. Hindahl (St. Johannes Hospital Dortmund, Germany); G. Ehninger, M. Schaich (Universitätsklinikum Dresden, Germany); A. Mackensen, S. Krause (Universitätsklinikum Erlangen, Germany); M. Geißler, J. Bauer (Klinikum Esslingen, Germany); H. Serve, C. Brandts (Universitätsklinikum Frankfurt/Main, Germany); M. Kiehl, W. Stein (Klinikum Frankfurt/Oder, Germany); H.-G. Höffkes, O. Ranze (Städtisches Klinikum Fulda, Germany); N. Schmitz, R. Stuhlmann (Asklepios Klinik St. Georg, Hamburg, Germany); H. Schmidt, K. Buhrmann (Kreiskrankenhaus Hameln, Germany); H.A. Dürk, D. Metzner (St. Marien-Hospital Hamm, Germany); A. D. Ho, A. Krämer (Universitätsklinikum Heidelberg, Germany); U. Kaiser, A. Bartholomäus (St. Bernward-Krankenhaus Hildesheim, Germany); A.A. Fauser, N. Basara (Klinik für Hämatologie/Onkologie und KMT, Idar-Oberstein, Germany); H. Link, S. Mahlmann (Westpfalzklinikum Kaiserslautern, Germany); M. Wolf, B. Ritter (Klinikum Kassel, Germany); L. Mantovani-Löffler, D. Kürschner (Städtisches Klinikum St. Georg Leipzig, Germany); T. Neuhaus, C. Hoffmann (St. Vincent Krankenhaus Limburg/Lahn, Germany); S. Fetscher, J. Schmielau (Sanakliniken Lübeck, Germany); H. Lehnert, S. Brüggemann (Universitätsklinikum Lübeck, Germany); A. Neubauer, K. Sohlbach (Universitätsklinikum Giessen und Marburg, Germany); E. Schleyer (Klinikum Merseburg, Germany); M. Griesshammer, H.-J. Tischler (Klinikum Minden, Germany); L. Lutz, M. Hentrich (Städtisches Krankenhaus München-Harlaching, Germany); W.E. Berdel, C. Müller-Tidow, (Universitätsklinikum Münster, Germany); H. Wandt, K. Schäfer-Eckart (Klinikum Nord, Nürnberg, Germany); A. Jakob, I. Dresel (Kreiskrankenhaus Offenburg, Germany); T. Gaska, E. Niemeyer (Brüderkrankenhaus Paderborn, Germany); T. Kozák, J. Vydra (University Hospital Praha, Czech Republic); A. Reichle, E. Holler (Universitätsklinikum Regensburg, Germany); F. Heits, A. Meinhardt (Diakonissen-Krankenhaus Rotenburg, Germany); T. Geer, I. Hrusovsky (Diakonie-Krankenhaus Schwäbisch Hall, Germany); S. Kanzler, H. H. Reinel (Lepoldina Krankenhaus Schweinfurt, Germany); E. Heidemann, J. Kaesberger (Diakonissenkrankenhaus Stuttgart, Germany); W.E. Aulitzky, L. Leimer (Robert-Bosch-Krankenhaus Stuttgart, Germany); M.R. Clemens, R. Mahlberg (Mutterhaus der Borromaerinnen Trier, Germany); N. Frickhofen, H.-G. Fuhr (Horst-Schmidt-Kliniken Wiesbaden, Germany); H. Einsele, M.-E. Goebeler (Universitätsklinikum Würzburg, Germany); M. Sandmann, G. Becker (Klinikum St. Antonius Wuppertal, Germany).