Key Points
Lyl1 is required for Lmo2-induced T-cell leukemia in mice, whereas Scl is dispensable.
LYL1 is required for growth of ETP-ALL cell lines.
Abstract
Lmo2 is an oncogenic transcription factor that is frequently overexpressed in T-cell acute lymphoblastic leukemia (T-ALL), including early T-cell precursor ALL (ETP-ALL) cases with poor prognosis. Lmo2 must be recruited to DNA by binding to the hematopoietic basic helix-loop-helix factors Scl/Tal1 or Lyl1. However, it is unknown which of these factors can mediate the leukemic activity of Lmo2. To address this, we have generated Lmo2-transgenic mice lacking either Scl or Lyl1 in the thymus. We show that although Scl is dispensable for Lmo2-driven leukemia, Lyl1 is critical for all oncogenic functions of Lmo2, including upregulation of a stem cell–like gene signature, aberrant self-renewal of thymocytes, and subsequent generation of T-cell leukemia. Lyl1 expression is restricted to preleukemic and leukemic stem cell populations in this model, providing a molecular explanation for the stage-specific expression of the Lmo2-induced gene expression program. Moreover, LMO2 and LYL1 are coexpressed in ETP-ALL patient samples, and LYL1 is required for growth of ETP-ALL cell lines. Thus, the LMO2-LYL1 interaction is a promising therapeutic target for inhibiting self-renewing cancer stem cells in T-ALL, including poor-prognosis ETP-ALL cases.
Introduction
Although intensive combinations of chemotherapeutic agents have improved the overall survival rate of T-cell acute lymphoblastic leukemia (T-ALL), the prognosis of patients who experience relapse remains poor because of outgrowth of resistant cells that arise from ancestral clones.1 One potential strategy to improve cure rates is to target the underlying primary genetic driver mutation that is common to all clones.
T-ALL can be subclassified based on the aberrant expression of transcription factor oncogenes.2-4 The largest of these comprises almost half of all T-ALL cases and is characterized by the overexpression of basic helix-loop-helix (bHLH) transcription factors such as SCL/TAL1 (hereafter termed “SCL”) or LYL1, along with their binding partners, the Lim domain–only proteins LMO1 or LMO2. Because these proteins interact to form transcription factor complexes, they are thought to act via a common mechanism to induce T-ALL.5 A further poor prognosis subtype of T-ALL can be identified by an immature immunophenotype with a gene expression signature similar to early T-cell precursors (termed “ETP-ALL”), including high expression of LYL1 but not SCL.4,6,7 The underlying genetic basis of ETP-ALL appears to be distinct from more mature T-ALL, with rearrangements activating transcription factors such as MEF2C and activating mutations of signaling pathways.4,8
LMO2 is overexpressed in >9% of T-ALL cases as a result of chromosomal abnormalities and is also highly expressed in ETP-ALL.2,4,6,9,10 In addition, LMO2 was overexpressed in 4 cases of T-ALL arising as a consequence of retroviral insertion mutagenesis after gene therapy for X-linked severe combined immunodeficiency (X-SCID).11,12 To understand the molecular basis of LMO2-induced T-ALL, the CD2-Lmo2 transgenic mouse model was developed.13 Overexpression of Lmo2 in the thymi of these mice results in T-cell lymphoma/leukemia, with an average latency of 10 months, highlighting the requirement for secondary mutations for disease. By using this model, we have shown that the primary effect of Lmo2 is to induce a stem cell–like phenotype including long-term self-renewal of thymocytes at the CD4-CD8-C25+CD44-(DN3) stage of T-cell development.10 These self-renewing immature T cells provide a pool of precancerous stem cells (pre-CSCs) that can acquire the additional mutations necessary for overt T-ALL. In addition, pre-CSCs in this model show remarkable resistance to conventional therapeutics such as γ-irradiation, suggesting that they may provide a source of disease relapse.10,14
During normal hematopoiesis, Lmo2 plays critical roles in hematopoietic stem cell (HSC) development, erythropoiesis, and angiogenesis but is redundant for T-cell development.15-18 The Lmo2 protein is comprised almost entirely of zinc-binding Lim domains that function to mediate protein-protein interactions.19 In the case of erythroid cells, the LMO2 complex has been well-characterized, where it comprises heterodimers of E2A and SCL, together with LDB1 and GATA1.20 This complex binds to a bipartite E-box-GATA DNA motif and interacts with transcriptional coactivator and repressor proteins to positively and negatively regulate transcription.21
Because of its inability to bind to DNA, the function of Lmo2 is thought to require its interaction with DNA-binding bHLH transcription factors. In addition to Scl, Lmo2 interacts with the related hematopoietic bHLH factor Lyl1.22 During early hematopoietic development, Lmo2 interacts with Scl, and both proteins are essential for this process.15,23-25 In contrast, Lyl1 is not required for hematopoietic development and cannot replace the function of Scl.26 During adult hematopoiesis, however, there is compensation between Scl and Lyl1 such that both proteins must be removed to ablate HSC function.27
In Lmo2-driven T-cell leukemia, the critical bHLH cofactor is currently unknown. For one, initial studies identified a multifactor DNA-binding complex comprising Lmo2 together with Scl.28 These proteins complex with E proteins (E47 or E12) and Ldb1 to bind to a bipartite E-box recognition motif. In addition, Scl can co-operate with Lmo2 in inducing T-cell leukemia in mouse models.29,30 However, gene expression analyses of Lmo2-transgenic thymocytes have demonstrated a gene expression profile that is similar to that of human immature T-ALL cases, including overexpression of Lyl1 but not Scl.10 Furthermore, microarray data of human ALL cases demonstrated increased expression of LMO2 in LYL1-expressing, immature T-ALL cases.2,4,6,10 To resolve this issue, we have used genetic ablation of either Scl or Lyl1 in an Lmo2-transgenic mouse model of T-cell leukemia to determine the relative contribution of these factors to Lmo2-driven thymocyte self-renewal and leukemogenesis. We demonstrate that Lyl1, but not Scl, is essential for Lmo2 to induce T-ALL in mice.
Materials and methods
Mice and poly(I:C) treatment
The CD2-Lmo2 transgenic,13 Scl knockout,24 Scl floxed,31 Lyl1 knockout,32 Mx-Cre transgenic,33 and ROSA26-YFP Cre-reporter mice34 have been described previously. All strains had been back-crossed to a C57BL/6 background. C57BL/6-Ly5.1 congenic recipient mice were obtained from the Walter and Eliza Hall Institute (WEHI). All animal experiments were approved by the animal ethics committees of the Faculty of Medicine, Dentistry and Health Sciences, University of Melbourne and the WEHI.
To induce the Mx1-Cre allele, 7-week-old mice were injected intraperitoneally 3 times at 48-hour intervals with 300 μg of polyinosinic-polycytidylic acid (poly(I:C)) (sodium salt; Sigma) dissolved in normal saline.
Flow cytometry
All antibodies used for this analysis were obtained from BD Pharmingen (San Diego, CA) or prepared by the WEHI monoclonal antibody facility: CD4 (GK 1.5), CD8 (clone 53-6.7), CD45.1 (A20), CD45.2 (104), Kit (CD117, 2B8), CD44 (IM7), CD25 (RM2-5), Mac1 (M1/70), Ter119 (Ly-76), and B220 (R3-6B2) as conjugates to FITC, PE, PE-Cy7, APC, Alexa700, and Biotin. For detection of biotinylated antibodies, streptavidin-PerCP-Cy5.5 (Pharmingen) was used. Data were acquired on FACSCalibur or Fortessa flow cytometers (BD Pharmingen). Sorting was performed on a FACSAria II Flow Cytometer (BD Pharmingen).
Southern blotting
Deletion of the Sclfl allele was determined by Southern blotting of EcoRI-digested genomic DNA using an Scl probe (probe B) that distinguishes floxed (fl), deleted (Δ), wild-type (+), and null (–) Scl alleles, as in Hall et al.31 HindIII-digested lambda DNA was used as size markers.
Transplantation studies
Thymocyte transplantation studies were performed as in McCormack et al.10 Briefly, C57BL/6-Ly5.1 congenic recipient mice were irradiated with 6.5 Gy from a 60Co γ source. Mice were then injected via the tail vein with thymocytes derived from one quarter of a thymus and maintained with water that contained antibiotics.
For tumor transplantations, recipient C57BL/6-Ly5.1 mice were irradiated as described before. Cells derived from Lmo2-induced thymomas were FACS-purified, then 3 × 105 (tumor 575) or 105 (tumor 580) of each population were injected via the tail vein.
Microarray analyses
See supplemental Methods.
GEO accession number: GSE49164
Quantitative polymerase chain reaction
See supplemental Methods.
shRNA lentiviral transduction of human cell lines
K562, Jurkat, PEER, and LOUCY cell lines were maintained in RPMI medium containing 10% fetal calf serum and 50 μM 2-mercaptoethanol. For lentiviral transduction, 105 cells were spinfected with MISSION shRNA lentiviral transduction particles (Sigma) according to the manufacturer’s protocol, at a multiplicity of infection of 1. The clones used were TRCN0000017538, TRCN0000017539, TRCN0000017540, TRCN0000017541, TRCN0000424749, and TRCN0000429914 (hereafter numbered according to the last 2 digits of the clone number). pLKO.1 (empty vector) particles were used as a control. Three days after infection, 0.5 μg/mL of Puromycin was added and the viable cell count was determined at various time points using Trucount beads (BD Pharmingen) and a FACScan flow cytometer (BD Pharmingen).
Results
Scl is dispensable for Lmo2-driven thymocyte self-renewal and leukemia
We have previously shown that overexpression of Lmo2 induces self-renewal of immature CD4-CD8 double-negative (DN) thymocytes. Given the recognized genetic and biochemical interaction of Lmo2 with Scl in T-ALL,28-30 we examined the expression of Scl and Lyl1 in DN thymocyte subsets of wild-type and Lmo2-transgenic mice. Expression of both Scl and Lyl1 were calculated according to known quantities of plasmid so that the relative amounts of Scl and Lyl1 mRNA could be compared. In wild-type cells, there was two- to threefold more Lyl1 than Scl mRNA in thymocytes, with both silenced by the DN4 stage (supplemental Figure 1). The presence of the Lmo2 transgene increased Lyl1 expression but not Scl in DN3 and DN4 thymocytes.
To examine the role of these bHLH factors in Lmo2-driven leukemia, we first used Scl conditional knockout mice to determine whether this factor is required for Lmo2-induced T-ALL. To delete Scl specifically in the T-cell lineage, we bred CD2-Lmo2 transgenic mice with mice containing conditionally targeted (Sclfl) and knockout Scl– alleles, along with the Lck-Cre transgene, to generate CD2-Lmo2;Lck-Cre;SclΔ/– mice (hereafter termed “Lmo2;Lck;SclΔ/– mice”). Deletion of Scl had no effect on T-cell development in mice that did not express the Lmo2 transgene, indicating that, like Lmo2,18 Scl is dispensable for this process (supplemental Figure 2).
CD2-Lmo2 transgenic mice exhibit thymocyte self-renewal from a young age, resulting in a progressive accumulation of immature CD4-CD8 DN thymocytes.10,13 We therefore examined the thymocyte profile of Lmo2;Lck;SclΔ/– mice at 6 months of age. As expected, CD2-Lmo2 transgenic mice showed accumulation of DN3 thymocytes (CD4–CD8–CD44–CD25+) with abnormally high levels of Kit, as seen previously.10 Absence of one or both alleles of Scl had no effect on the increase of DN thymocytes seen in Lmo2-transgenic mice (Figure 1A-B). In addition, the reduction in thymic cellularity caused by overexpression of Lmo2 was still apparent in the absence of Scl (Figure 1C). To confirm that the Scl allele was deleted in the thymocytes of these mice, genomic DNA was prepared from thymocytes derived from 6-month-old Lmo2;Lck;SclΔ/– mice. Southern blotting confirmed that the Scl allele was deleted in the thymocytes of these mice, but not other hematopoietic lineages (Figure 1D). Thus, Scl is dispensable for the accumulation of DN3 thymocytes caused by Lmo2 overexpression.
Scl is not required for Lmo2-induced thymocyte self-renewal. (A) Accumulation of immature (CD4-CD8 DN) thymocytes in Lmo2;Lck;SclΔ/– mice. Representative fluorescence-activated cell sorting analysis of total (top panel) and DN thymocytes (center and lower panels) from 2-month-old mice of the indicated genotypes. wt, wild-type; Tg, transgenic). (B) Analysis of the percentage of DN thymocytes in preleukemic mice at 6 months of age. (C) Thymic cellularity of preleukemic mice at 6 months of age. Data represent mean +SD of 3 to 5 mice per group. (D) Scl is effectively deleted in Lmo2;Lck;SclΔ/– thymocytes. Genomic DNA was extracted from the bone marrow (BM) and thymus (THY) of 6-month-old Lmo2;Lck;SclΔ/– mice and used in Southern hybridization to reveal null (–), floxed (fl), and deleted (Δ) Scl alleles. %DN indicates the percentage of DN thymocytes in the thymi analyzed. (E) Scl is not required for long-term engraftment of Lmo2-transgenic thymocytes. Thymocytes were isolated from 2-month-old mice of the indicated genotypes, and thymocytes equivalent to one-quarter of a thymus were injected into sublethally irradiated (6.5 Gy) Ly5.1 recipients. Three weeks later, the proportion of donor thymocytes was determined by flow cytometry. Points represent individual recipient mice, and unique symbols denote separate experiments. (F) Donor thymocytes from recipient mice as in (D) were gated and analyzed for CD4 and CD8 expression by flow cytometry.
Scl is not required for Lmo2-induced thymocyte self-renewal. (A) Accumulation of immature (CD4-CD8 DN) thymocytes in Lmo2;Lck;SclΔ/– mice. Representative fluorescence-activated cell sorting analysis of total (top panel) and DN thymocytes (center and lower panels) from 2-month-old mice of the indicated genotypes. wt, wild-type; Tg, transgenic). (B) Analysis of the percentage of DN thymocytes in preleukemic mice at 6 months of age. (C) Thymic cellularity of preleukemic mice at 6 months of age. Data represent mean +SD of 3 to 5 mice per group. (D) Scl is effectively deleted in Lmo2;Lck;SclΔ/– thymocytes. Genomic DNA was extracted from the bone marrow (BM) and thymus (THY) of 6-month-old Lmo2;Lck;SclΔ/– mice and used in Southern hybridization to reveal null (–), floxed (fl), and deleted (Δ) Scl alleles. %DN indicates the percentage of DN thymocytes in the thymi analyzed. (E) Scl is not required for long-term engraftment of Lmo2-transgenic thymocytes. Thymocytes were isolated from 2-month-old mice of the indicated genotypes, and thymocytes equivalent to one-quarter of a thymus were injected into sublethally irradiated (6.5 Gy) Ly5.1 recipients. Three weeks later, the proportion of donor thymocytes was determined by flow cytometry. Points represent individual recipient mice, and unique symbols denote separate experiments. (F) Donor thymocytes from recipient mice as in (D) were gated and analyzed for CD4 and CD8 expression by flow cytometry.
The ability of Lmo2-transgenic thymocytes to repopulate the thymus in transplantation assays provides evidence that Lmo2 can induce self-renewal of T cells.10 To determine whether Lmo2-induced thymocyte self-renewal requires Scl, Lmo2;Lck;SclΔ/Δ thymocytes were injected into sublethally irradiated mice, and the extent of thymic engraftment was determined 3 weeks later. As expected, normal thymocytes had no repopulating capacity (Figure 1E). In contrast, transplant of Lmo2;Lck;SclD/D thymocytes demonstrated long-term engraftment capacity similar to Lmo2;Lck;SclΔ/+ controls (Figure 1E). Analysis of the T-cell differentiation profile of donor thymocytes demonstrated all thymocyte subsets including double-positive (DP) and single-positive thymocytes (Figure 1F). Thus, self-renewal of Lmo2 transgenic thymocytes does not require Scl.
A cohort of Lmo2;Lck;SclΔ/– mice were monitored long term to determine the requirement of Scl for Lmo2-driven T-cell leukemia. The latency of T-ALL in Lmo2;Lck;SclΔ/– mice was slightly delayed compared with Lmo2;Lck;SclΔ/+ controls, although this did not reach statistical significance (Figure 2A; P = .06). Southern blot analysis of thymic tumors arising in Lmo2;Lck;SclΔ/– mice confirmed complete deletion of the Sclfl allele, as well as the presence of Scl-deleted tumor cells infiltrating the bone marrow and spleen (Figure 2B). Together, these data indicate that Scl is dispensable for Lmo2-induced thymocyte self-renewal and T-cell leukemogenesis.
Scl is not required for Lmo2-driven leukemogenesis. (A) Kaplan-Meier survival curve of Lmo2-transgenic mice of the indicated genotypes. (P = .06 between SclΔ/+ and SclΔ/– using the log-rank [Mantel-Cox] test). (B) Deletion of Scl in hematopoietic tissues from a leukemic Lmo2;Lck;Scl mouse. Genomic DNA was extracted from the bone marrow (BM), spleen (SPL), and thymus (THY) of a leukemic Lmo2;Lck;SclΔ/– mouse and used in Southern hybridization to reveal null (–), floxed (fl), and deleted (Δ) Scl alleles.
Scl is not required for Lmo2-driven leukemogenesis. (A) Kaplan-Meier survival curve of Lmo2-transgenic mice of the indicated genotypes. (P = .06 between SclΔ/+ and SclΔ/– using the log-rank [Mantel-Cox] test). (B) Deletion of Scl in hematopoietic tissues from a leukemic Lmo2;Lck;Scl mouse. Genomic DNA was extracted from the bone marrow (BM), spleen (SPL), and thymus (THY) of a leukemic Lmo2;Lck;SclΔ/– mouse and used in Southern hybridization to reveal null (–), floxed (fl), and deleted (Δ) Scl alleles.
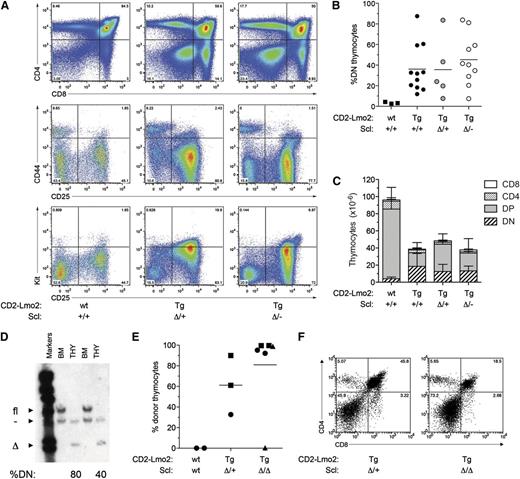
Lyl1 is essential for Lmo2-induced T-cell differentiation block, thymocyte self-renewal, and leukemia
The increased expression of Lyl1 in Lmo2-transgenic thymocytes suggested that Lyl1 may be the critical bHLH cofactor for Lmo2 rather than Scl (supplemental Figure 1).10 To determine the importance of Lyl1 in Lmo2-induced T-ALL, Lmo2 transgenic mice were bred with Lyl1-knockout (Lyl1−/−) mice32 to generate Lmo2 transgenic mice heterozygous (CD2-Lmo2;Lyl1+/−) or null (CD2-Lmo2;Lyl1+/−) for Lyl1. Analysis of Lyl1−/− mice not expressing the Lmo2 transgene revealed a decrease in early thymic progenitors (ETPs) and DN2 cells, as recently reported (supplemental Figure 3).35 However, the number of DN3 thymocytes, the stage at which Lmo2 induces self-renewal,10 was only slightly reduced in Lyl1−/− mice. Strikingly, however, the absence of Lyl1 completely prevented the increased proportion of DN thymocytes seen in CD2-Lmo2 transgenic mice (Figure 3A-B). Moreover, thymus size, which was reduced in Lmo2-transgenic mice, was restored to normal in the absence of Lyl1 (Figure 3C). In contrast, heterozygosity of Lyl1 had no significant effect on the proportion of DN thymocytes or thymic cellularity in CD2-Lmo2 transgenic mice.
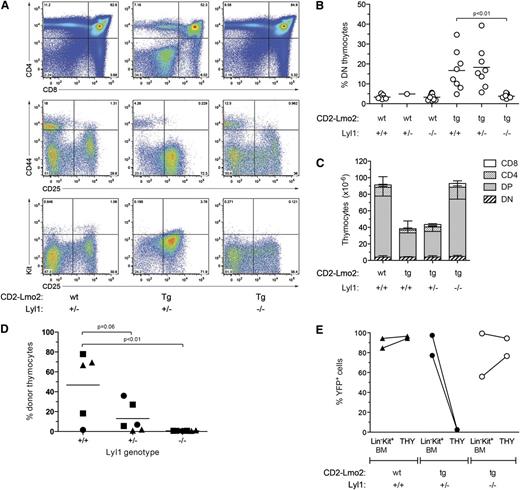
Lyl1 is required for Lmo2-induced differentiation block and thymocyte self-renewal. (A) Absence of DN thymocyte accumulation in Lmo2;Lyl1−/− mice. Flow cytometric profiles of thymocytes from 2-month-old mice of the indicated genotypes are shown. (B) Cumulative analysis of the percentage of immature (DN) thymocytes in CD2-Lmo2;Lyl1−/− mice at 2 to 3 months of age. P values were calculated using Student t test. (C) Normalization of thymocyte cellularity in Lmo2;Lyl1−/− mice. Numbers of various thymocyte subsets in 2-month-old mice of the indicated genotypes (n = 3 per genotype) were determined by flow cytometry. Numbers are given as mean +SD. (D) Lyl1 is required for long-term transplantation of Lmo2-transgenic thymocytes. Thymocytes were isolated from 2-month-old, Lmo2-transgenic mice of the indicated Lyl1 genotypes, and thymocytes derived from one-quarter of a thymus were injected into sublethally irradiated (6.5 Gy) Ly5.1 recipients. Three weeks later, the proportion of donor thymocytes was determined by flow cytometry. P values were determined using Student t test. (E) Loss of Lyl1 restores progenitor importation into the thymus of Lmo2-transgenic mice. Mx;YFP mice of the indicated genotypes were injected with poly(I:C) at 7 weeks of age. Three weeks later, the percentages of YFP+ BM progenitors (Lin–Kit+ BM) and thymocytes (THY) were determined.
Lyl1 is required for Lmo2-induced differentiation block and thymocyte self-renewal. (A) Absence of DN thymocyte accumulation in Lmo2;Lyl1−/− mice. Flow cytometric profiles of thymocytes from 2-month-old mice of the indicated genotypes are shown. (B) Cumulative analysis of the percentage of immature (DN) thymocytes in CD2-Lmo2;Lyl1−/− mice at 2 to 3 months of age. P values were calculated using Student t test. (C) Normalization of thymocyte cellularity in Lmo2;Lyl1−/− mice. Numbers of various thymocyte subsets in 2-month-old mice of the indicated genotypes (n = 3 per genotype) were determined by flow cytometry. Numbers are given as mean +SD. (D) Lyl1 is required for long-term transplantation of Lmo2-transgenic thymocytes. Thymocytes were isolated from 2-month-old, Lmo2-transgenic mice of the indicated Lyl1 genotypes, and thymocytes derived from one-quarter of a thymus were injected into sublethally irradiated (6.5 Gy) Ly5.1 recipients. Three weeks later, the proportion of donor thymocytes was determined by flow cytometry. P values were determined using Student t test. (E) Loss of Lyl1 restores progenitor importation into the thymus of Lmo2-transgenic mice. Mx;YFP mice of the indicated genotypes were injected with poly(I:C) at 7 weeks of age. Three weeks later, the percentages of YFP+ BM progenitors (Lin–Kit+ BM) and thymocytes (THY) were determined.
The restoration of thymic differentiation and cellularity in the absence of Lyl1 suggested that Lyl1 was required for Lmo2-driven thymocyte self-renewal. To examine this directly, Lmo2;Lyl1−/− thymocytes were transplanted into sublethally irradiated recipients, and the proportion of donor thymocytes was measured 4 weeks later. In contrast to control CD2-Lmo2 thymocytes, donor CD2-Lmo2;Lyl1−/− thymocytes were incapable of long-term engraftment (Figure 3D). In addition, Lmo2 transgenic thymocytes lacking a single allele of Lyl1 (Lmo2;Lyl1+/−) showed a trend toward reduced engraftment (P = .06, Student t test).
The lack of long-term engraftment capacity in CD2-Lmo2;Lyl1−/− thymocytes could be a result of impaired homing of donor thymocytes rather than the loss of self-renewal capacity. To address this possibility, we used a lineage-tracing assay that we have previously developed, which can track thymic turnover from the bone marrow without the need for irradiation or transplantation.10 Briefly, this assay uses poly(I:C) treatment of Mx-Cre transgenic mice carrying a ROSA-EYFP Cre-reporter allele, which activates expression of YFP in HSCs but not thymocytes. The kinetics of thymic turnover from labeled (YFP+) bone marrow–derived progenitors can subsequently be measured. Using this assay, we have shown that the presence of self-renewing thymocytes in CD2-Lmo2 transgenic mice blocks thymic turnover from the bone marrow at a young age, and therefore Lmo2-transgenic thymocytes remain YFP-negative.10 To examine whether loss of Lyl1 restores thymocyte turnover in CD2-Lmo2 transgenic mice, we generated CD2-Lmo2;Mx-Cre;ROSA-EYFP mice that were heterozygous or nullizygous for Lyl1. These mice were treated with poly(I:C) to induce the Mx-Cre transgene, and 4 weeks later the proportion of YFP+ thymocytes was determined. As previously reported, >90% of thymocytes in wild-type mice became YFP+ 4 weeks after administration of poly(I:C) (Figure 3E). In contrast, Lmo2-transgenic thymocytes heterozygous for Lyl1 remained YFP-negative, indicating the presence of self-renewing thymocytes that block thymocyte turnover from the bone marrow.10 Strikingly, however, thymic turnover in CD2-Lmo2;Lyl1−/− mice was completely restored, with levels of YFP+ thymocytes similar to that of bone marrow progenitors (Figure 3E). Together with the thymocyte transplantation experiment, these data indicate that Lyl1 is required for Lmo2-induced thymocyte self-renewal in vivo.
Our previous gene expression analyses of Lmo2-transgenic DN thymocytes demonstrated that aberrant self-renewal capacity was associated with reactivation/maintenance of an HSC-like transcriptional program.10 To determine whether this expression program requires Lyl1, we performed microarray analyses on DN thymocytes derived from wild-type, Lmo2, and Lmo2;Lyl1−/− mice. This revealed a complete normalization of Lmo2-upregulated genes in CD2-Lmo2;Lyl1−/− mice (Figure 4A). Notably, the expression of Hhex, a direct target gene of Lmo2 that can itself induce thymocyte self-renewal, was restored to normal levels after loss of Lyl1.10,36 In addition to upregulating a stem cell–like transcriptional program in thymocytes, overexpression of Lmo2 represses a number of important T-cell developmental genes including the pre–T cell receptor α subunit (Ptcra). Repression of these genes was also dependent on the presence of Lyl1 (Figure 4B). Thus, Lyl1 is essential for all transcriptional effects of overexpressed Lmo2 in T cells.
Lyl1 is required for Lmo2-driven leukemogenesis. (A) Normalization of gene expression in Lmo2;Lyl1−/− thymocytes. A heat map shows the expression levels of the top 40 Lmo2-upregulated genes in wild-type (wt), Lmo2-transgenic (Tg), and Lmo2;Lyl1−/− DN thymocytes. (B) A heat map shows the expression levels of the top 40 Lmo2-downregulated genes in DN thymocytes of mice of the indicated genotypes. Labels are as in (A). (C) Kaplan-Meier survival curve of mice of the indicated genotypes. P < .001 between Lmo2;Lyl1+/− and Lmo2;Lyl−/− (log-rank [Mantel-Cox] test).
Lyl1 is required for Lmo2-driven leukemogenesis. (A) Normalization of gene expression in Lmo2;Lyl1−/− thymocytes. A heat map shows the expression levels of the top 40 Lmo2-upregulated genes in wild-type (wt), Lmo2-transgenic (Tg), and Lmo2;Lyl1−/− DN thymocytes. (B) A heat map shows the expression levels of the top 40 Lmo2-downregulated genes in DN thymocytes of mice of the indicated genotypes. Labels are as in (A). (C) Kaplan-Meier survival curve of mice of the indicated genotypes. P < .001 between Lmo2;Lyl1+/− and Lmo2;Lyl−/− (log-rank [Mantel-Cox] test).
We next monitored cohorts of Lmo2 transgenic mice of various Lyl1 genotypes for the development of leukemia. Lmo2 transgenic, Lyl1-heterozygous mice developed leukemia at a similar rate to that previously seen for Lmo2-transgenic mice (Figure 4C).13 However, only 2 of 19 Lmo2-transgenic mice lacking Lyl1 developed leukemia during the 16-month observation period. Genotyping of these tumors confirmed the Lyl1−/− genotype. Surprisingly, 2 of 7 Lyl1−/− mice without the Lmo2 transgene also developed leukemia, suggesting that the leukemias occurring in the Lmo2; Lyl1−/− mice were independent of the Lmo2 transgene. Together, these results indicate that Lyl1 is required for the oncogenic effect of Lmo2 in T cells.
Lyl1 expression is restricted to stem cell populations in preleukemia and leukemia
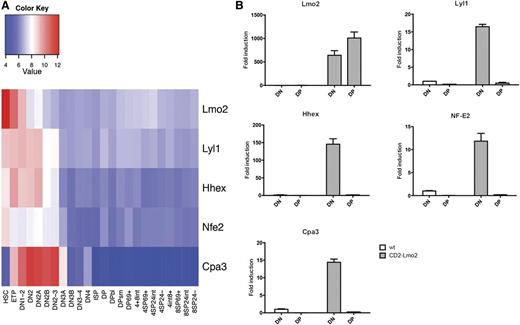
We have previously demonstrated that Lmo2-mediated thymocyte self-renewal is restricted to immature thymocytes within the DN3 fraction.10 Our finding that Lyl1 is a critical cofactor for Lmo2 in developing thymocytes suggests that stage-specific Lyl1 expression may affect the expression of Lmo2-induced gene expression programs and thereby modulate its oncogenic activity. To examine this, we assessed the normal expression pattern of genes upregulated by Lmo2 during T-cell development using microarray gene expression data.37 Similar to Lyl1 expression (supplemental Figure 1), several genes upregulated in Lmo2-transgenic thymocytes (Hhex, Nfe2, and Cpa3) are normally downregulated at the DN2 stage of T-cell development (Figure 5A). Because mature (DP) Lmo2-transgenic thymocytes do not have self-renewal capacity, we asked whether this may be due to loss of Lyl1 expression. Therefore, we examined the expression of Lmo2, Lyl1, Hhex, Nfe2, and Cpa3 in sorted DP cells from wild-type and Lmo2-transgenic mice. We found that although Lmo2 expression is maintained in DP thymocytes of Lmo2-transgenic mice, Lyl1 expression is downregulated more than 30-fold compared with DN cells (Figure 5B). Similarly, expression of Hhex, Nfe2, and Cpa3 was also restricted to DN thymocytes in Lmo2-transgenic mice (Figure 5B). These data suggest that the stage-specific expression of Lyl1 restricts the transcriptional effects of overexpressed Lmo2 to immature T cells, providing a molecular explanation for our previous findings that self-renewal potential is restricted to DN thymocytes.10
Downregulation of Lyl1 during differentiation of wild-type and Lmo2-transgenic thymocytes. (A) Expression of Lmo2, Lyl1, and their target genes during normal T-cell differentiation. Data from the Immgen Database37 were used to determine the expression levels of the indicated genes during normal αβ T-cell differentiation. ETP, early thymocyte precursor; ISP, CD8+ intermediate single-positive cell. (B) Downregulation of Lmo2 target genes during T-cell differentiation in Lmo2-transgenic mice. DN and DP thymocytes were sorted from 8-week-old mice of the indicated genotypes and used to generate cDNA, which was amplified by real-time PCR using primers specific for the indicated genes. Data are represented as mean +SD of triplicate values and are representative of 3 separate experiments.
Downregulation of Lyl1 during differentiation of wild-type and Lmo2-transgenic thymocytes. (A) Expression of Lmo2, Lyl1, and their target genes during normal T-cell differentiation. Data from the Immgen Database37 were used to determine the expression levels of the indicated genes during normal αβ T-cell differentiation. ETP, early thymocyte precursor; ISP, CD8+ intermediate single-positive cell. (B) Downregulation of Lmo2 target genes during T-cell differentiation in Lmo2-transgenic mice. DN and DP thymocytes were sorted from 8-week-old mice of the indicated genotypes and used to generate cDNA, which was amplified by real-time PCR using primers specific for the indicated genes. Data are represented as mean +SD of triplicate values and are representative of 3 separate experiments.
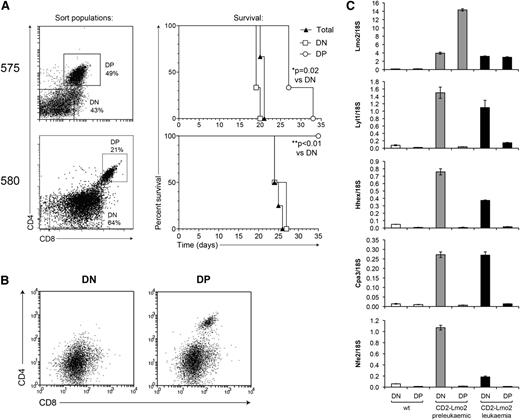
Next we examined whether Lyl1 expression is also restricted to cancer stem cells in overt T-cell leukemia. To address this, we fractionated tumors from Lmo2-transgenic mice into DN and DP populations to evaluate their potential to induce tumors in secondary recipients. Transplant of DN cells from tumor 575 generated leukemia more rapidly than did equivalent numbers of DP cells, and in the case of tumor 580, leukemia-initiating cells were restricted to the DN fraction (Figure 6A). Moreover, secondary leukemias developing from DP cells of tumor 575 displayed a predominant DN surface phenotype, suggesting that the transplanted DP population was either contaminated with DN cells or capable of de-differentiation (Figure 6B). DN cells in secondary tumors expressed CD44 and CD25, but not Kit, indicating that they are reminiscent of DN2-DN3 thymocytes (supplemental Figure 4). These results indicate that CSCs in overt Lmo2-induced T-cell leukemia are restricted to the immature (DN2-DN3) population of tumor cells, consistent with other reports that used similar mouse models.38,39 We then assessed the expression of Lmo2 and Lyl1 and their targets in CSC-containing DN tumor cells vs more differentiated DP cells using quantitative polymerase chain reaction (PCR). Similar to preleukemic thymocytes, expression of Lyl1, Hhex, Cpa3, and Nfe2 were downregulated in DP leukemic thymocytes despite ongoing expression of Lmo2 (Figure 6C). Together, these data suggest that Lyl1 expression acts to restrict the expression of the Lmo2-induced HSC-like expression program to stem cell populations in both preleukemia and overt leukemia.
The Lmo2-associated gene expression program is restricted to CSCs. (A) Tumor samples derived from Lmo2-transgenic mice were fractionated into the indicated populations, and 3 × 105 cells of each population were injected into irradiated congenic (Ly5.1) recipients. The survival curves of mice receiving the indicated tumor cell populations are shown on the right. (B) FACS profile of secondary tumors from mice receiving the indicated primary tumor populations from mouse 575. (C) DN and DP cells were sorted from 2-month-old wild-type (wt) or Lmo2-transgenic mice, or from Lmo2-transgenic tumor 575 (as in A), and the expression levels of the indicated genes were determined by real-time PCR.
The Lmo2-associated gene expression program is restricted to CSCs. (A) Tumor samples derived from Lmo2-transgenic mice were fractionated into the indicated populations, and 3 × 105 cells of each population were injected into irradiated congenic (Ly5.1) recipients. The survival curves of mice receiving the indicated tumor cell populations are shown on the right. (B) FACS profile of secondary tumors from mice receiving the indicated primary tumor populations from mouse 575. (C) DN and DP cells were sorted from 2-month-old wild-type (wt) or Lmo2-transgenic mice, or from Lmo2-transgenic tumor 575 (as in A), and the expression levels of the indicated genes were determined by real-time PCR.
LYL1 is expressed in ETP-ALL and required for growth of ETP-ALL cell lines
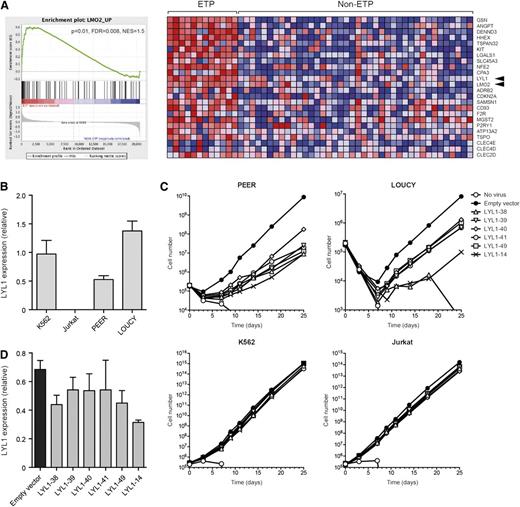
Finally, we sought to examine the functional relevance of the Lmo2-Lyl1 interaction in human T-ALL. We have previously shown that the expression of LMO2 and several LMO2-associated genes is associated with the LYL1-expressing subset of T-ALL.10 This subset was proposed to relate to the recently described ETP ALL subset, which exhibits immature features and poor prognosis.4,7,8 To determine whether LMO2 and LYL1 are coexpressed in ETP-ALL, we performed gene set enrichment analysis using our described Lmo2-upregulated gene set10 and published microarray data from ETP and non-ETP T-ALL patient samples.8 We found a strong association between Lmo2-upregulated genes and ETP-ALL (Figure 7A). Notably, LMO2 and LYL1 were coexpressed in 11 of 12 ETP patient samples (92%), with the remaining sample expressing high levels of LYL1 but intermediate levels of LMO2. This high expression occurred despite none of these ETP-ALL patients carrying chromosomal abnormalities in either gene.8 In contrast, only 7 of 40 (18%) non-ETP T-ALL cases coexpressed LMO2 and LYL1. Thus, the gene expression profile of Lmo2-induced pre-CSCs is highly similar to that of human ETP-ALL patients.
Lmo2 and LYL1 are coexpressed in ETP-ALL, and LYL1 is required for maintenance of ETP-ALL lines. (A) Gene set enrichment analysis of Lmo2-upregulated genes in ETP vs non-ETP T-ALL. Lmo2-upregulated genes10 were compared with microarray data from patient samples of ETP and non-ETP T-ALL.8 The enrichment plot (left) shows skewing to the left, indicating association with the ETP subclass of T-ALL. The heat map (right) shows the “leading edge” genes that are most associated with ETP-ALL. Arrowheads indicate LMO2 and LYL1. FDR, false discovery rate; NES, normalized enrichment score. (B) Real-time PCR showing expression of LYL1 in the indicated cell lines. (C) Growth curves of the indicated cell lines infected with the indicated LYL1 knockdown lentiviral vectors and selected in Puromycin. (D) Real-time PCR showing expression of LYL1 in K562 cells infected with the indicated LYL1-knockdown lentiviral vectors and selected in Puromycin.
Lmo2 and LYL1 are coexpressed in ETP-ALL, and LYL1 is required for maintenance of ETP-ALL lines. (A) Gene set enrichment analysis of Lmo2-upregulated genes in ETP vs non-ETP T-ALL. Lmo2-upregulated genes10 were compared with microarray data from patient samples of ETP and non-ETP T-ALL.8 The enrichment plot (left) shows skewing to the left, indicating association with the ETP subclass of T-ALL. The heat map (right) shows the “leading edge” genes that are most associated with ETP-ALL. Arrowheads indicate LMO2 and LYL1. FDR, false discovery rate; NES, normalized enrichment score. (B) Real-time PCR showing expression of LYL1 in the indicated cell lines. (C) Growth curves of the indicated cell lines infected with the indicated LYL1 knockdown lentiviral vectors and selected in Puromycin. (D) Real-time PCR showing expression of LYL1 in K562 cells infected with the indicated LYL1-knockdown lentiviral vectors and selected in Puromycin.
To test the functional relevance of LMO2/LYL1 expression in ETP-ALL, we obtained 2 ETP-ALL cell lines, PEER and LOUCY, both of which express high levels of LMO2 and LYL1.40 Real-time PCR confirmed a high expression of LYL1 in both lines, but not the mature T-ALL cell line Jurkat (Figure 7B).40 We then infected each of these lines with 6 separate LYL1-knockdown lentiviral vectors. This demonstrated a profound reduction in growth of the ETP-ALL lines (PEER and LOUCY), but no growth inhibition of Jurkat cells or of K562, an erythroleukemic cell line that expresses LYL1 (Figure 7A,C). Notably, the 2 lentiviral vectors that gave the greatest knockdown of LYL1 in K562 cells (LYL1-38 and -14) also gave the greatest growth inhibition of the ETP-ALL lines. Thus, LYL1 is specifically required for growth of ETP-ALL cell lines.
Discussion
Transcription factors play central roles in both normal tissue development and cancer but have been difficult to target therapeutically. One approach to tackling this problem is to identify crucial protein-protein interactions that can be exploited to interfere with oncogenic function. The hematopoietic bHLH transcription factors Scl and Lyl1 are almost identical within their bHLH domains and, as such, Lmo2 can interact with either of these proteins but not a variety of other bHLH proteins.22 Here we have examined which of these proteins are required for Lmo2-induced T-cell leukemogenesis. Our finding that Lmo2 requires Lyl1, but not Scl, to elicit leukemia identifies a protein-protein interaction that may be exploited therapeutically to inhibit Lmo2 function.
Our data demonstrate that Scl is dispensable for tumorigenesis in Lmo2-transgenic mice. There was an apparent increase in engraftment of Lmo2 preleukemic cells lacking Scl; however, this did not correlate with an earlier onset of leukemia (Figure 1). Rather, leukemia in Scl-deleted mice was slightly delayed, suggesting that the low level of Scl expression in Lmo2-transgenic thymocytes may help to facilitate tumorigenesis (Figure 2A).
In contrast to adult HSCs, where Scl and Lyl1 are functionally redundant,27 we found that Lyl1 is essential for all aspects of Lmo2-induced leukemia, including differentiation arrest, thymocyte self-renewal, and the aberrant transcriptional effects of Lmo2, including the upregulation of HSC-associated genes and concomitant downregulation of T-cell developmental genes. Accordingly, downregulation of Lyl1 at the DN-to-DP transition coincides with reduced expression of Lmo2 target genes, including Nfe2, Hhex, and Cpa3. This may provide a molecular explanation for the observation that Lmo2 overexpression is not oncogenic in mature T cells.41 The concurrent expression of these genes during normal T-cell development (Figure 6) implies that they are part of an immature gene expression program that must be extinguished for T-cell development to proceed, as has been proposed previously.42
Because we have shown that the transcriptional profile of Lmo2-induced pre-CSCs is similar to that of human ETP-ALL (Figure 7A), we propose that Lmo2 transgenic mice provide a model for this subtype, which has a poor prognosis with standard therapies.2,4,6-8 In the case of Lmo2-indcued T-ALL, the increased expression of Lyl1 is likely to be direct because LMO2 binds and activates the LYL1 promoter, forming a positive regulatory network with the downstream target gene Hhex.40 A similar positive regulatory network may also underpin MEF2C-induced ETP-ALL, where MEF2C binds to the LMO2 promoter and activates the expression of both LMO2 and LYL1.4 Accordingly, we have shown that LYL1 expression is required for growth of ETP-ALL cell lines (Figure 7C). Thus, expression of LMO2 and LYL1 appears to be a driving factor underpinning ETP-ALL, and inhibition of this interaction is a promising therapeutic approach to improving the outcome of this poor-prognosis subtype. Notably, inhibitors of Lmo2 have previously been shown to inhibit tumor transplantation in an Lck-Lmo2 mouse model, indicating a requirement for this factor in tumor propagation in vivo.43-45
We found a low frequency of T-cell leukemia in Lyl1 knockout mice, both in the presence and the absence of the Lmo2 transgene. The mechanism of leukemia in these mice is unknown, but it may relate to the defects in lymphoid specification and maintenance of ETPs in Lyl1 knockout mice (supplemental Figure 3).35 Despite a reduction in DN2 cells in Lyl1−/− mice, numbers of DN3 cells were only slightly reduced, suggesting that compensatory mechanisms increase proliferation of DN3 thymocytes in these mice, which may provide the opportunity for leukemogenic mutations to occur.
In summary, our data demonstrate that Lyl1 is required for the oncogenic effects of Lmo2 in developing T cells, and it is required for growth of ETP-ALL cell lines. These results warrant further investigation of the therapeutic potential of inhibition of the Lmo2-Lyl1 interaction as a means to inhibit self-renewing cancer stem cells in LMO2-expressing T-cell leukemias, in particular the ETP-ALL subtype, which shows poor response to current treatments.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lan Ta and WEHI Bioservices for excellent animal husbandry and Dina Stockwell for technical assistance. We thank Dr. John Pimanda for the LOUCY cell line.
This work was supported by Project Grants (628386 and 1003391 [M.P.M., D.J.C.]) and an Independent Research Institute Infrastructure Support (IRIIS) Scheme from the Australian National Health and Medical Research Council (NHMRC), grants-in-aid from the Cancer Council of Victoria and the Leukaemia Foundation of Australia (M.P.M.), a Future Fellowship from the Australian Research Council (M.P.M), a Senior Medical Research Fellowship from the Sylvia and Charles Viertel Foundation (D.J.C), and a Victorian State Government Operational Infrastructure Grant. THR is supported by Programme Grants from Leukaemia and Lymphoma Research and the Medical Research Council.
Authorship
Contribution: M.P.M and D.J.C. designed research, analyzed data, and wrote the manuscript; M.P.M., B.J.S, J.T.J., C.N., W.S., N.J.S., and C.S.T. performed research and analyzed data; and T.H.R. designed research and provided reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew P. McCormack, Cancer and Haematology Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville VIC 3052, Victoria, Australia; e-mail: mccormack@wehi.edu.au.

![Figure 2. Scl is not required for Lmo2-driven leukemogenesis. (A) Kaplan-Meier survival curve of Lmo2-transgenic mice of the indicated genotypes. (P = .06 between SclΔ/+ and SclΔ/– using the log-rank [Mantel-Cox] test). (B) Deletion of Scl in hematopoietic tissues from a leukemic Lmo2;Lck;Scl mouse. Genomic DNA was extracted from the bone marrow (BM), spleen (SPL), and thymus (THY) of a leukemic Lmo2;Lck;SclΔ/– mouse and used in Southern hybridization to reveal null (–), floxed (fl), and deleted (Δ) Scl alleles.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/12/10.1182_blood-2012-09-458570/4/m_2093f2.jpeg?Expires=1768015336&Signature=i1NRYxHLYjXUR6jA8kFTNZlvTsHze-XBJaaFal1qjp86XzJgjkrV38reN0Dw7h5Zyr5ykIXMm6g5OLms9eSEyRx6aF9FPMyvd6BTbbmXrhzpqty0huStuP-128mIczNwwZOIH3OWHAVr~gqL2NIpsq3uNC48GRhhqLxzyGwAvgS77CvnVhU~knIZq5ELYAvaDC6PQkyMPOnGC-~RWOyCjsGREl7vmn3CnZJ56CB2Io5niNLIncwIHhVP3kY7E3XcvXWL~Y0Mka2gFKqnlOwYya0fXDPtamwAKeas0sf-tsnha-ERJK84n9lKl-yLH2FNLdDTtdJD02flp-BrGEFCGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Lyl1 is required for Lmo2-driven leukemogenesis. (A) Normalization of gene expression in Lmo2;Lyl1−/− thymocytes. A heat map shows the expression levels of the top 40 Lmo2-upregulated genes in wild-type (wt), Lmo2-transgenic (Tg), and Lmo2;Lyl1−/− DN thymocytes. (B) A heat map shows the expression levels of the top 40 Lmo2-downregulated genes in DN thymocytes of mice of the indicated genotypes. Labels are as in (A). (C) Kaplan-Meier survival curve of mice of the indicated genotypes. P < .001 between Lmo2;Lyl1+/− and Lmo2;Lyl−/− (log-rank [Mantel-Cox] test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/12/10.1182_blood-2012-09-458570/4/m_2093f4.jpeg?Expires=1768015336&Signature=VRTngIlQ03r7XATgegphAgn-QriHLpD5aw9zOzcyxPa-F0FwXlxFsokoFvCIYRQiNflvyBbBHJVTwfg70LjkRAg3nRXrkVPbn2z31BGVLkuNlxqKnD1OCbtu2AZtXGuQtKDh-zw-7A9El6jOEF3zcn9QVBCReo9a3Cc1Zn0hHJLY6UNrzUj34WxhjTy8zmpp-Ap0GVnN43Ajo9TcPqH-d-hA3j2TobdEAZhi6Kjlul-wFMoVhshlD8ce2YJOmebRUuXf7sdVt-E3EdHrU4-dvcdwAD87Ux6bfCcHGhmZetakKSYMDGzOGn66gQESDGAKiYTaQxMReyH~4Vd8-lvs6A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)