Key Points
Somatic copy number alterations of miRNA genes are uncommon in de novo and secondary AML.
MIR223 silencing in AML occurs through both genetic and epigenetic mechanisms.
Abstract
Altered microRNA (miRNA) expression is frequently observed in acute myelogenous leukemia (AML) and has been implicated in leukemic transformation. Whether somatic copy number alterations (CNAs) are a frequent cause of altered miRNA gene expression is largely unknown. Herein, we used comparative genomic hybridization with a custom high-resolution miRNA-centric array and/or whole-genome sequence data to identify somatic CNAs involving miRNA genes in 113 cases of AML, including 50 cases of de novo AML, 18 cases of relapsed AML, 15 cases of secondary AML following myelodysplastic syndrome, and 30 cases of therapy-related AML. We identified a total of 48 somatic miRNA gene-containing CNAs that were not identified by routine cytogenetics in 20 patients (18%). All these CNAs also included one or more protein coding genes. We identified a single case with a hemizygous deletion of MIR223, resulting in the complete loss of miR-223 expression. Three other cases of AML were identified with very low to absent miR-223 expression, an miRNA gene known to play a key role in myelopoiesis. However, in these cases, no somatic genetic alteration of MIR223 was identified, suggesting epigenetic silencing. These data show that somatic CNAs specifically targeting miRNA genes are uncommon in AML.
Introduction
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression posttranscriptionally by binding to target messenger RNAs (mRNAs).1 Although miRNAs are frequently dysregulated in acute myelogenous leukemia (AML),2-9 the mechanism of dysregulation remains poorly understood. It is known that the majority of human miRNA genes are present in fragile sites and genomic regions frequently altered in cancer.10 Point mutations of miRNA genes appear to be rare in human cancers. While single nucleotide polymorphisms (SNPs) in miRNAs that affect expression have been reported,11,12 there are only rare examples of recurring somatic point mutations in miRNA genes in human cancer.13,14 Conversely, somatic copy number alterations (CNAs) that include miRNA genes have been reported in several human cancers.15-18 However, whether miRNA genes are frequently and specifically targeted in AML by deletion or amplification is largely unknown. To address this issue, we performed a comprehensive analysis of somatic CNAs involving miRNA genes in 113 cases of AML (50 cases of de novo AML, 18 cases of relapsed AML, 15 cases of secondary AML following myelodysplastic syndrome, and 30 cases of therapy-related AML [t-AML]) by using custom miRNA-specific, high-resolution array-based comparative genomic hybridization (aCGH) and whole-genome sequence data.
Methods
Human subjects
All AML samples were obtained from a study at Washington University to identify genetic factors contributing to AML initiation and progression. Approval for these studies was obtained from the Washington University institutional review board. After obtaining written informed consent for the patients in accordance with the Declaration of Helsinki, a bone marrow sample and a 6-mm punch biopsy of skin (for analysis of matched normal cells) were obtained.
aCGH
A custom high-resolution aCGH platform (3×720K array; NimbleGen, Madison, WI) was generated to interrogate CNAs of all known miRNA genes at the time these studies were performed (835 miRNAs [miRBase, version 14.0] for the 30 t-AML samples and 1027 miRNAs [miRBase, version 15.0] for the 18 relapsed AML samples) and 44 miRNA processing genes (Table 1). Each gene and 40 kb of its flanking genome were interrogated with densely tiled probes at either 30 to 40 bp (miRNA genes) or 80 bp (miRNA processing genes). This array also contained dense tiling of probes designed to interrogate 170 DNA repair genes. In addition, probes uniformly spaced throughout the genome at approximately 8600-bp intervals were included. Two micrograms of genomic DNA from unfractionated bone marrow (tumor) and paired normal tissue (skin) was fragmented, labeled, and hybridized to the array as previously described.19 Log2 ratios of fluorescent intensity for tumor/skin were generated for each probe. Abnormal segments (ie, putative regions of CNAs) were identified by using segmentation algorithms from NimbleGen (segments) and Partek (segmentation). Segments generated by segmentation algorithms were prioritized on the basis of the number of probes and the log2 ratio of each segment (score = log10 [number of probes per segment] × log2 ratio) and manually reviewed, as previously described.19 To identify CNAs within miRNA genes and miRNA processing gene loci, plots of log2 values for each probe spanning the locus with 0.5 to 5 Mb flanking DNA were manually reviewed by 4 independent reviewers. Next, we collapsed contiguous segments generated by segmentation algorithms and identified boundaries by using segment boundaries and manual review. For 18 of the 30 t-AML patients, an independent iScan platform was available, and it confirmed 100% of the aCGH calls.
Analysis of whole-genome sequencing data
We recently reported the sequence of 50 de novo AML genomes20 and 15 genomes of patients with secondary AML following myelodysplastic syndrome.21,22 The sequence data were analyzed to identify potential somatic CNAs as previously described.20 However, there is a high false-positive rate with CNAs identified in this fashion.21 Thus, we also performed aCGH by using the Affymetrix 6.0 SNP array to independently call somatic CNAs in all cases. We included for further analysis only those CNAs that were identified by both platforms.
Real time RT-PCR
Total RNA was reverse transcribed by using the TaqMan microRNA Reverse Transcription Kit per manufacturer’s instructions (Applied Biosystems). Real time reverse transcription-polymerase chain reaction (RT-PCR) for the indicated miRNA and RNU48 (as a control) were performed by using the relevant TaqMan MicroRNA assay.
Quantitative genomic PCR
Quantitative PCR was performed by using SYBR Green Master Mix (Applied Biosystems) and 50 ng of genomic DNA. PCR primers were designed to amplify MIR223 and MIR181b. MIR181b was included as a diploid gene copy number control, since no somatic CNAs of this gene were identified in any of the samples. MIR223 primers were 5′-CTTTACCTGCTTATCTTCAGGATCTCT-3′ and 5′-CGTACGCGCCCCCATCAGCACTCT-3′. MIR181b primers were 5′-GTCTCCCATCCCCTTCAGAT-3′ and 5′-TTTGCCTTTTCTAAAACATGCTC-3′. Technical triplicates were performed for each sample.
Results and discussion
A total of 113 patients with AML were studied, including 50 cases of de novo AML (Table 2), 18 cases of relapsed de novo AML (Table 2), 15 cases of secondary AML following myelodysplastic syndrome (Table 3), and 30 cases of t-AML (Table 4). The median age of the de novo AML patients was 54.5 years (range, 21 to 82 years), and the median blast percentage was 75% (range, 35% to 100%). A normal karyotype was identified in 37 (74%) of 50 patients. The median age of the relapsed AML patients was 57.5 years (range, 24 to 77 years). The median blast percentage was 59% (range, 12% to 95%). A normal karyotype was identified in 6 (40%) of 15 patients with relapsed AML. The patients with secondary AML were older, with a median age of 66 years (range, 26 to 77 years). The median time to progression from myelodysplastic syndrome to AML was 400 days (range, 28 to 1751 days), and the median blast percentage in the bone marrow was 43% (range, 21% to 89%). A normal karyotype was identified in 43% of cases, and abnormalities involving chromosome 5 or 7 were observed in 43%. The median age of patients with t-AML was 59 years (range, 26 to 80 years). Twelve of the t-AML patients (40%) were treated for breast cancer, 6 (20%) for non-Hodgkin lymphoma, 2 (6.7%) for Hodgkin lymphoma, 2 (6.7%) for multiple myeloma, and 8 (20%) for other diseases. Most of the t-AML patients (76.7%) were treated for their primary cancer with a combination of chemotherapy that included topoisomerase inhibitors and/or alkylating agents. The median blast percentage in the bone marrow was 76% (range, 31% to 95%). Cytogenetic analysis revealed −5/−5q and/or −7 in seven patients (23%), translocations involving chromosome 11q23 (MLL gene rearrangement) in 6 patients (20.0%), and a normal karyotype in 6 patients (20%).
We interrogated paired tumor/normal samples for somatic CNAs by using aCGH or whole-genome sequencing data. The t-AML and relapsed AML cases were analyzed by using a custom CGH array that contained densely spaced oligomers (every 30 to 40 bp spacing) for all miRNA genes that were identified in miRBase at the time this study was performed (835 miRNAs in miRBase, version 14.0, were included in the arrays for the 30 t-AML samples and 1027 miRNAs in miRBase, version 15.0, for the 18 relapsed de novo AML samples). A total of 40 kb of genomic DNA flanking the miRNA precursor gene was targeted. We also included probes for 44 genes involved in miRNA processing (Table 1). In each case, genomic DNA isolated from a skin biopsy was used to distinguish inherited CNAs from somatic CNAs. To call a somatic CNA, we required that a minimum of 25 contiguous probes show differential hybridization. Thus, for miRNA genes, we theoretically should be able to identify somatic CNAs of approximately 1 kb. A total of 64 CNAs that were not apparent by routine cytogenetics were identified in 14 patients (all with t-AML). CNAs were judged to be cytogenetically apparent if any part of the contiguous segment was contained within a chromosomal loss, gain, or interstitial chromosomal deletion identified by routine metaphase cytogenetics. For interstitial deletions, coordinates of the cytogenetic banding were estimated by using National Center for Biotechnology Information (NCBI) Map Viewer, Build 36. Twenty-six of these somatic CNAs, identified in 11 of the 48 patients, contained one or more miRNA genes (Table 5). No cytogenetically unapparent somatic CNAs involving miRNA processing genes were identified in any case.
To expand our analysis, we next analyzed whole-genome sequencing and aCGH data for 50 cases of de novo AML and 15 cases of secondary AML to identify somatic CNAs. For these samples, the Affymetrix 6.0 SNP array was used. We required that the CNAs be identified by both whole-genome sequencing and by aCGH. Given the lower probe density of the Affymetrix 6.0 SNP array, we estimated that the lower size limit of somatic CNA detection for this approach was approximately 18 kb. Four somatic CNAs involving miRNA genes were identified in 4 de novo AML patients, all with a normal karyotype (Table 5). In the secondary AML cases, we identified 18 somatic CNAs in 5 patients, only one of which had a normal karyotype. In total, we identified cytogenetically unapparent somatic CNAs involving miRNA genes in 18% of patients with AML. In AML with a normal karyotype, somatic CNAs involving miRNA genes were identified in only 5 (9.1%) of 55 cases. The most common recurring somatic CNA (present in 3 cases of AML) is an approximately 1.3-Mb deletion at 17q11.2, which includes MIR4733, MIR4724, MIR193a, MIR4725, and MIR365b (Table 5). However, as is the case for all of the somatic CNAs identified in this study, the 17q11.2 CNA includes several protein coding genes.
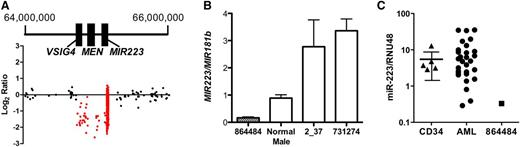
The smallest somatic CNA identified in this study is a 429-kb deletion on chromosome X that includes MIR223 and two other genes, MSN and VSIG4 (Figure 1A). It occurred in a male patient with t-AML with complex cytogenetics (Table 4, unique patient number [UPN] 864484). Quantitative PCR of genomic DNA isolated from the bone marrow of this patient confirmed a hemizygous deletion of MIR223 (Figure 1B). As expected, the hemizygous deletion of MIR223 in this patient resulted in the complete loss of miR-223 expression (Figure 1C). miR-223 is one of the most highly expressed miRNAs in human CD34+ cells,23 and its expression increases with myeloid differentiation.24 Accordingly, miR-223 has been implicated in granulocytic differentiation. Fazi et al24 showed that enforced expression of miR-223 in acute promyelocytic leukemic cells induces granulocytic differentiation. Conversely, loss of Mir223 is associated with a myeloproliferative-like phenotype in mice.25
Hemizygous loss of MIR-223 in a patient with AML. (A) Log2 ratio dot plots of paired tumor and normal DNA from patient UPN 864484 analyzed by using the custom CGH array. A discrete deletion of approximately 429 kb on chromosome X is depicted. Genomic coordinates are based on NCIBI36/HG18 assembly. (B) Quantitative PCR for MIR223 and MIR181b (control gene) was performed by using genomic DNA from the indicated source. Shown is the ratio of MIR223 to MIR181b signal. Data represent the mean ± standard error of the mean of triplicate measurements. (C) miR-223 expression relative to RNU48 is shown for CD34+ cells isolated from healthy donors (CD34) and leukemic bone marrow from patient UPN 864484 or 28 other patients with AML. The 90% confidence interval is shown for CD34+ cells.
Hemizygous loss of MIR-223 in a patient with AML. (A) Log2 ratio dot plots of paired tumor and normal DNA from patient UPN 864484 analyzed by using the custom CGH array. A discrete deletion of approximately 429 kb on chromosome X is depicted. Genomic coordinates are based on NCIBI36/HG18 assembly. (B) Quantitative PCR for MIR223 and MIR181b (control gene) was performed by using genomic DNA from the indicated source. Shown is the ratio of MIR223 to MIR181b signal. Data represent the mean ± standard error of the mean of triplicate measurements. (C) miR-223 expression relative to RNU48 is shown for CD34+ cells isolated from healthy donors (CD34) and leukemic bone marrow from patient UPN 864484 or 28 other patients with AML. The 90% confidence interval is shown for CD34+ cells.
To determine whether loss of miR-223 expression was a common occurrence in AML, we performed real-time RT-PCR on bone marrow RNA from an additional 28 cases of AML and from CD34+ cells isolated from 5 healthy donors (Figure 1C). We identified three cases in which miR-223 expression was below the 90% confidence interval based on normal CD34+ cells. Two of these samples (UPN 2_37 and 731274) were from male patients. Quantitative PCR performed on genomic DNA isolated from their leukemic bone marrow showed no deletion of MIR223 (Figure 1B). The third sample with very low miR-223 expression (UPN 189941) was from a female patient. The sequence of her leukemic genome was recently reported and revealed no point mutation or CNA of MIR223.26 Thus, in all of these cases, an epigenetic mechanism is the likely cause of miR-223 silencing. Indeed, UPN 2_37 (a 46-year-old male with M1 AML) had a t(8;21) translocation producing the AML-ETO fusion oncogene, which has been shown to epigenetically silence MIR223.14,27 Our data suggest that the deletion of MIR223 represents another, albeit uncommon, mechanism to decrease miR-223 expression in AML.
Although miRNAs are frequently dysregulated in AML, it appears that genetic alterations in miRNA are relatively rare. Results from whole genome sequencing of 24 cases of de novo AML identified recurring point mutations in a single miRNA gene.21 Specifically, point mutations in MIR142 were identified in 2% of cases of de novo AML. Our study suggests that small somatic CNAs involving miRNA genes that are not apparent by standard cytogenetics are uncommon. Thus, it appears that epigenetic, rather than genetic, mechanisms are responsible for most cases of miRNA dysregulation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Translational Research Program Award from the Leukemia & Lymphoma Society (D.C.L.) and by grants RC2 CA1455073 (D.C.L.) and PO1-CA101937 (T.J.L.) from the National Institutes of Health.
Authorship
Contribution: G.R., M.A.J., M.J.W., and D.C.L. designed the custom comparative genomic hybridization array; G.R., M.A.J., J.S., R.E.D.J.P., D.S., M.T., A.H.G., and M.J.W. contributed to data analysis; T.J.L. provided crucial reagents (acute myelogenous leukemia samples); and D.C.L. was responsible for the overall design and analysis of all studies and edited the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Division of Oncology, Washington University School of Medicine, Campus Box 8007, 660 South Euclid Ave, St. Louis, MO 63110; e-mail: dlink@dom.wustl.edu.
References
Author notes
G.R. and M.A.J. contributed equally to this study.