Key Points
The cellular ligand of the NKp44L is a novel isoform of the mixed-lineage leukemia-5 protein.
NKp44L is not expressed on healthy cells, but on tumor and transformed cells, rendering them more sensitive for the NK cytotoxicity.
Abstract
With an array of activating and inhibitory receptors, natural killer (NK) cells are involved in the eradication of infected, transformed, and tumor cells. NKp44 is a member of the natural cytotoxicity receptor family, which is exclusively expressed on activated NK cells. Here, we identify natural cytotoxicity receptor NKp44 (NKp44L), a novel isoform of the mixed-lineage leukemia-5 protein, as a cellular ligand for NKp44. Unlike the other MLL family members, NKp44L is excluded from the nucleus, but expressed at the cell-surface level; its subcellular localization is being associated with the presence of a specific C-terminal motif. Strikingly, NKp44L has not been detected on circulating cells isolated from healthy individuals, but it is expressed on a large panel of the tumor and transformed cells. The sharply decreased NK lysis activity induced by anti-NKp44L antibodies directly demonstrates the role of NKp44L in cytotoxicity. Taken together, these results show that NKp44L could be critical for NK cell-mediated innate immunity. The identification and cellular distribution of NKp44L highlight the role of this self-molecule as a danger signal to alert the NK cell network.
Introduction
Natural killer (NK) cells are critical components of the innate immune response, comprising the first line of defense against a variety of tumors and microbial pathogens before any sensitization.1-3 An 11-year follow-up study clearly showed that the cytolytic activity of NK cells is associated with reduced cancer risk.4 The effector function of NK cells is regulated by a dynamic interplay between activating and inhibitory signals that are delivered by the interaction of various ligands and receptors at the surface of NK cells and able to sense cell stress, malignancy, and infection.5 There is compelling evidence that activating signals are required for NK cells to trigger abnormal cells. These major activating receptors include NKG2D, DNAM-1, 2B4, and the natural cytotoxicity receptors (NCR).6 The NCR family comprises NKp46, NKp44, and NKp30. Both resting and activated NK cells express NKp30 and NKp46, whereas NKp44 expression is limited to activated NK cells. NKp44 is also expressed by certain decidual NK cells,7 NK-22 cells from mucosal-associated lymphoid tissues,8 and a subset of tonsil interferon-producing cells.9 The de novo expression of NKp44 may play a role in the marked increase of cytolytic activity displayed by activated NK cells, whereas the lysis of some target cells is only marginally NKp44-dependent, as demonstrated by the lack of significant interference with cytotoxicity by antibody-mediated blocking of this molecule.10,11
The interaction between NK cells and their targets depends on the expression of ligands specific for each NCR, but determining these has proved difficult. The first NCR ligands identified were of viral origin.12-16 Until now, the only NK cellular ligands clearly identified have been for NKp30, including the nuclear HLA-B-associated transcript-3 protein that plays a role in NK/dendritic cell crosstalk,17 and the cell-surface B7-H6 protein only detected on tumor cell lines of various origins.18,19
The cellular ligand of NKp44, called natural cytotoxicity receptor NKp44 (NKp44L), is expressed on CD4+ T cells during HIV infection,20,21 and its expression is correlated with the decline of the CD4+ T-cell count, both ex vivo in HIV-infected patients,20 and in vivo in SHIV-infected macaques.22
To better understand the relations between NKp44 and the pathophysiology of both infectious diseases and malignancies, we undertook studies to identify and characterize its cellular ligand (NKp44L). Here, we found that NKp44L is an isoform of mixed-lineage leukemia-5 (MLL5), located in the human chromosome band 7q22. NKp44L was not detectable at a steady state in the normal tissues but was present in a broad range of hematopoietic and nonhematopoietic tumor and transformed cells, where it could trigger autologous NK cells. Altogether, our results provide new insights into the role of the cellular ligands for activating NK receptors as danger signals to alert the NK cell network.
Methods
The cDNA library construction and yeast two-hybrid screening
A two-hybrid cDNA library was constructed from poly(A+) messenger RNA (mRNA) of Jurkat cells (selected for insert sizes between 500 and 4000 bp) (supplemental Methods, available on the Blood Web site). This library directionally inserted downstream of the LexA transcriptional activation domain in the pYESTrp vector (Invitrogen) was transformed into the yeast strain EGY191/pSH18-34 containing the bait vector with the extracellular domain of NKp44. Colonies that grew on selective medium were assayed for β-galactosidase activity by a filter assay.23 Plasmids isolated from primary positive yeasts were retransformed into yeasts in conjunction with either NKp44 bait vector or various control baits (Jun, NKp46, or NKp30) (supplemental Methods).
A 5′-rapid amplification of cDNA ends, northern blot, and RT-PCR analysis
A 5′-rapid amplification of cDNA ends was performed from mRNAs that had been reverse-transcribed with the SuperScript preamplification system (Life Technologies). Polymerase chain reaction (PCR) was performed with the PfuTurbo DNA polymerase (Stratagene). All amplified products were then sequenced.
Northern blot and reverse transcription-polymerase chain reaction (RT-PCR) assays are described in the supplemental Methods section.
NKp44L mammalian vectors and production of stable transfectants
The DNA sequence coding for the full-length NKp44L sequence was amplified from polyA+ RNA purified from Jurkat cells by PCR and then cloned in a cloning vector. C-terminal NKp44L truncation was performed by PCR mutagenesis to obtain the NKp44L sequence deleted of the 3′ terminal 36 bp, corresponding with the 21spe exon (44L-Δ21spe). The PCR products were cut and subcloned into a pFLAG-CMV4 plasmid (Sigma-Aldrich) and their DNA inserts were checked with an automatic sequencer ABI 3100 (Applied Biosystems) to check the DNA sequence integrity.
EL4 cells were stably transfected with the pFLAG-CMV4 constructs or the pFLAG-CMV4-BAP control plasmid by electroporation (250 V, 500 μF). Cells were then selected in the presence of G418 (Clontech) for at least 20 days. At least two different cell populations were selected and tested by flow cytometry and functional assays.
Flow cytometry
Fluorescence-activated cell sorter analysis was performed on freshly harvested cells, as described.20 Cells were incubated with either 20 μg/mL of anti-NKp44L mAbs (#7.1 immunoglobulin M [IgM]) (supplemental Methods)20 or 10 μg/mL of NKp30-Ig, NKp44-Ig, or NKp46-Ig fusion proteins (R&D Systems). Recombinant human IgG Fcγ-fragment-specific (R&D Systems), or mouse IgM isotype control (MOPC-104E; Sigma-Aldrich) served as negative controls. Cells were then incubated with rat anti-mouse IgM Ab (1/50) or Fcγ-fragment-specific conjugated affinity-purified F(ab′)2 fragments of goat anti-human IgG (1/50; Jackson ImmunoResearch Laboratory). For intracellular staining, cells were permeabilized with Cytofix/Cytoperm kit (Beckton Dickinson). Analysis was performed on FACSCalibur (Becton Dickinson) or Navios (Coulter) flow cytometers and analyzed with FlowJo software (Tree Star, Inc.).
Western blot and immunoprecipitation analysis
Immunoprecipitation experiments, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed, as described.24 Cells were lysed with NET buffer (150 mM NaCl, 50 mM Tris pH 7.4, 5 mM EDTA) containing 1% Tx-100, and protease inhibitor cocktails (Thermo Scientific), and subsequently centrifuged. Biotinylation of cell surface proteins was performed on cells incubated with 0.25 mg/mL EZ-link sulfo-NHS-LC-biotin (Pierce). The free biotin was removed, and the cells were then lysed with NET buffer.
For the immunoprecipitation experiments, lysates were incubated with rabbit polyclonal anti-MLL5 Ab (ab75339; Abcam), IgG1-Fc, NKp44-Ig, anti-actin (Sigma-Aldrich) coupled to Protein A/G UltraLink resin. Mouse IgM (Abcam) or anti-NKp44L mAb were coupled to Protein L agarose resin for immunoprecipitation (Thermo Scientific). Fusion proteins or mAb coupled resins were incubated with 500 µg of lysate overnight at 4°C. After washing, the immune complexes were eluted in a reducing sample buffer containing sodium dodecyl sulfate.
For western blot analysis, immune complexes or cell lysates were denatured in sample buffer, resolved on 8% SDS-PAGE (Bio-Rad) and transferred onto nitrocellulose membrane (Amersham). Membrane was blocked with 1% gelatin in phosphate-buffered saline 0.1% Tween 20, incubated with 2 µg/mL anti-NKp44L mAb, NKp44-Ig, or horseradish peroxidase-streptavidin (1/4000), or anti-actin (1/1000; Cell Signaling Technology), and then stained with secondary mAb coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratory.). Membranes were analyzed with an Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence
HeLa cells were cultured on sterilized glass coverslips at the bottom of 12-well microplates, whereas stably transfected EL4 cells were seeded on microscope slides by cytospin centrifugation (Shandon Cytospin). Cells were fixed with acetone and then stained with 10 µg/mL anti-NKp44L mAb, NKp44-Ig fusion protein, or rabbit polyclonal anti-MLL5 Ab (1/150). Cells were then incubated with Alexa Fluor 488-conjugated species-specific secondary antibodies (Molecular Probes/Invitrogen) for NKp44L and MLL5 detection or with DyLight 488-conjugated goat anti-human Fcγ-specific IgG (Jackson ImmunoResearch Laboratory) to reveal the NKp44-Ig and IgG1 Fc fusion proteins. Rabbit polyclonal anti-calnexin CT Ab (1/500) (SPA-860; Stressgen) was revealed by Alexa Fluor 555-conjugated goat anti-rabbit IgG (Molecular Probes). Cell coverslips were mounted onto glass slides in fluoromount G (Southern Biotechnology). Images were acquired with Apotome Zeiss and Axiovision 4.8 software.
Isolation of primary NK cells and NK cytotoxicity assays
NK cells from healthy donors (Etablissement Français du Sang) were negatively purified with the MACS NK-cell isolation kit (Miltenyi Biotec), according to the manufacturer’s recommendations. Purified NK cells were cultured in RPMI 1640 Glutamax medium (Invitrogen), supplemented with 1000 units interleukin 2 (IL-2) (Proleukin, Roche) for 10 days. The purity of these preparations was evaluated by flow cytometry in the presence of anti-NKp44 mAb. The cytolytic activity of NK cells was assayed in standard 4-hour 51Cr-release assays, as described.20 In the experiments including mAbs or isotype controls, these were added to the final concentration of 20 μg/mL.
Bioinformatic analysis
All sequences were analyzed with EMBL-EBI (www.ebi.ac.uk) and Expasy bioinformatics Resource Portal (www.expasy.org), which contain all available gene and protein prediction sequences from the TREMBL, Swiss-Protein, GenPept, ENSEMBL, GeneSeqP, PIR, PDB, and BLAST databases. The specific motif encoded in each protein was delineated with the Expasy database.
The study was approved by the Ile-de-France VI French Ethics Committee (Comité de Protection des Personnes). Informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Results
Identification of NKp44L by screening interaction with the extracellular domain of NKp44
Using the NCR-Ig fusion proteins obtained on fusion of each of the NKp30, NKp44, and NKp46 extracellular domains with the constant region of human IgG, we first confirmed the presence of NCR ligands at the cell surface of different transformed or tumor cell lines (supplemental Figure 1).25
Applying the yeast two-hybrid system to identify the protein interacting with the extracellular domain of NKp44, we constructed a bait that encoded a fusion protein containing the complete sequence of the extracellular part of NKp44 and used it to screen a cDNA library of Jurkat cells positive for NKp44L (supplemental Figure 1). Ten positive clones, isolated from 4 independent screenings, were grown on leucine-deficient (Leu−) medium. They activated LexA-dependent lacZ gene expression in the presence of the NKp44 bait plasmid, but not alone, or with irrelevant control baits encoding the protein Fos, and the extracellular domains of NKp46 and NKp30 (supplemental Methods). Sequencing of these 10 LacZ+Leu+ clones revealed that all contained inserts encoding a single sequence localized on human chromosome 7q22 (GenBank accession no. NT_007933.15). This sequence was 725 to 843 nucleotides in length, depending on the clone, and contained an open reading frame (ORF) of 129 to 170 amino acids, a stop codon (TAG), and a 3′ untranslated region (UTR), up to a polyadenylation site for certain clones (supplemental Table 1).
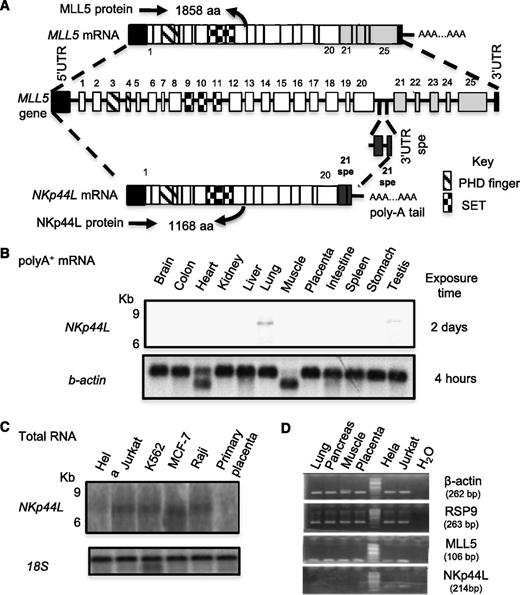
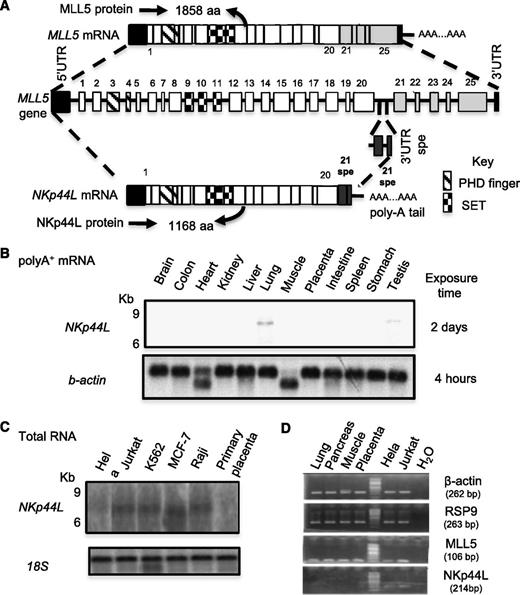
The sequence in 5′ was determined by performing sequential RACE on mRNA extracted from Jurkat cells. This 4034-bp cDNA fragment, designated NKp44L contains a 200-bp 5′UTR, a 3504-bp complete ORF, and a 3′ UTR of 330 nucleotides, up to the polyadenylation signal AATAAAA (GenBank accession no. JQ809698). Sequencing and BLAST analysis revealed that this fragment partially matched the gene encoding MLL5, spanning approximately 100 Kb of genomic DNA. Supplemental Table 2 describes its exon/intron structure. The first 20 exons are identical to those of the MLL5 gene, which contains 25 exons,26 but lacks the exon 21spe, which is specific to NKp44L (Figure 1A). This specific 21spe fragment was previously notified in the GenBank database (accession no. AAR13894.1). The ORF of NKp44L predicts a protein of 1168 amino acids that both contains PHD (Plant homeodomain)-zinc finger, and Suvar3-9, Enhancer of zeste, Trithorax motifs (Figure 1A; supplemental Figure 2).
Gene organization and mRNA expression of NKp44L. (A) Schematic representation of the NKp44L gene organization compared with MLL5. Exons are represented as bars and are numbered. Exons 21 to 25 are specific to MLL5, whereas 21spe exon is specific to NKp44L. Exon 21spe (spe, specific) and the following 3′UTR spe are localized in the intron following the exon 20 of MLL5. The PHD zinc-finger domain is indicated by hatched bars and the Suvar3-9, Enhancer of zeste, Trithorax (SET) domain by checkerboard bars. (B) The mRNA expression of NKp44L in normal tissues. Northern blot analysis of normal tissue poly(A)+ RNA was performed with a DNA probe from the specific NKp44L sequence after exposure times of 2 days at −80°C (top panel). The bottom panel shows hybridization with a control β-actin probe after a standard 4-hour exposure. (C) RNA expression of NKp44L in tumor cell lines (HeLa, Jurkat, MCF-7, and Raji) and primary placenta tissue. Northern blot analysis of total RNA was performed with a DNA probe from the specific NKp44L sequence (top panel). The bottom panel shows hybridization with a control 18S ribosomal RNA probe. (D) RT-PCR analysis of NKp44L and MLL5 from complementary DNA of healthy tissues (lung, pancreas, muscle and placenta), HeLa, and Jurkat tumor cells. β-actin and RSP9 were used as controls. The sized of each amplified fragment is noted.
Gene organization and mRNA expression of NKp44L. (A) Schematic representation of the NKp44L gene organization compared with MLL5. Exons are represented as bars and are numbered. Exons 21 to 25 are specific to MLL5, whereas 21spe exon is specific to NKp44L. Exon 21spe (spe, specific) and the following 3′UTR spe are localized in the intron following the exon 20 of MLL5. The PHD zinc-finger domain is indicated by hatched bars and the Suvar3-9, Enhancer of zeste, Trithorax (SET) domain by checkerboard bars. (B) The mRNA expression of NKp44L in normal tissues. Northern blot analysis of normal tissue poly(A)+ RNA was performed with a DNA probe from the specific NKp44L sequence after exposure times of 2 days at −80°C (top panel). The bottom panel shows hybridization with a control β-actin probe after a standard 4-hour exposure. (C) RNA expression of NKp44L in tumor cell lines (HeLa, Jurkat, MCF-7, and Raji) and primary placenta tissue. Northern blot analysis of total RNA was performed with a DNA probe from the specific NKp44L sequence (top panel). The bottom panel shows hybridization with a control 18S ribosomal RNA probe. (D) RT-PCR analysis of NKp44L and MLL5 from complementary DNA of healthy tissues (lung, pancreas, muscle and placenta), HeLa, and Jurkat tumor cells. β-actin and RSP9 were used as controls. The sized of each amplified fragment is noted.
Tissue expression of NKp44L transcripts
We then evaluated tissue expression of NKp44L by northern blot analysis with polyA+ mRNAs from a variety of normal tissues. Using a specific NKp44L probe including the sequence corresponding to the exon 21spe, not present in MLL5, we failed to detect NKp44L transcripts in any of the tissue samples tested, except very low expression from lung and testis (Figure 1B). High expression of the transcripts, however, is detected in total RNA extracted from several tumor cell lines (Figure 1C). RT-PCR analysis shows that NKp44L and MLL5 have 2 distinct profiles of expression; MLL5 is expressed in all tested healthy and tumor cells, whereas NKp44L is mainly expressed in tumor cell lines (Figure 1D).
Biochemical characterization of the NKp44L protein
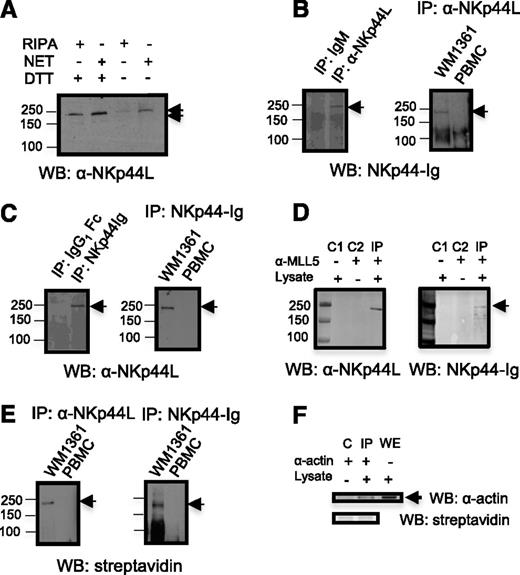
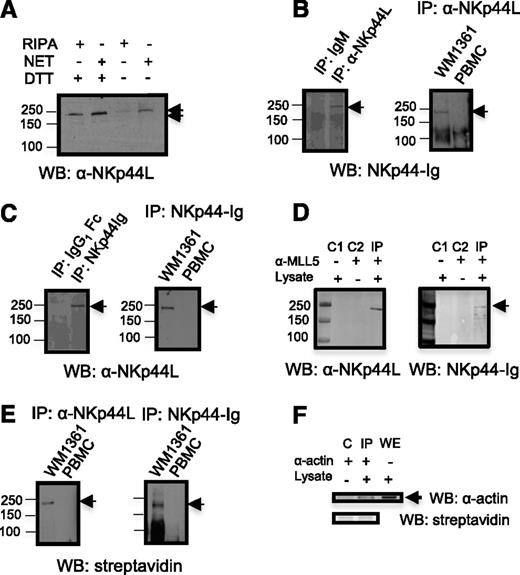
To further characterize NKp44L, direct western blot analysis of Jurkat cell lysate was performed in both nonreduced and dithiothreitol-reduced conditions with anti-NKp44L mAb, and revealed a shift of the unique band molecular-weight of approximately 250 KDa, consistent with treatment by a reducing agent that breaks the disulfide bridges (Figure 2A). More importantly, Figure 2B shows a similar band after immunoprecipitation of WM1361 cell lysate with anti-NKp44L mAb followed by immunoblotting with NKp44-Ig fusion protein, or after its immunoprecipitation with NKp44-Ig, then immunoblotting with anti-NKp44L mAb (Figure 2C). In contrast, no band was detected with lysate of peripheral blood mononuclear cells (PBMCs) from healthy donors (Figures 2B-C). This supports the specificity of the anti-NKp44L mAb for recognizing a cellular ligand of NKp44, also detected with the NKp44-Ig fusion protein. Furthermore, immunoprecipitation of WM1361 cell lysate with an anti-MLL5 Ab specific for the N-terminal domain of MLL5, expected to be present in NKp44L, also revealed a band around 250 kDa, reacting with both anti-NKp44L mAb or the NKp44-Ig fusion protein (Figure 2D). Similar data were observed with cell lysate from MT2 cells (data not shown). Notably, immunoprecipitation with anti-NKp44L mAb or the NKp44-Ig fusion protein of cell surface biotinylated WM1361 cells, followed by immunoblotting with horseradish peroxidase-streptavidin, also revealed the 250 kDa protein. Again, this band was not detected at the surface of biotinylated PBMCs from healthy donors (Figures 2E-F).
NKp44L immune-detection. (A) Direct western blot in the presence of nonreduced or reduced lysates. Proteins from Jurkat cells were extracted with radioimmunoprecipitation assay (RIPA) or NET buffer, treated or untreated with DTT, resolved on an 8% SDS-PAGE and then immunoblotted with anti-NKp44L mAb (αNKp44L). (B-C) Cell lysates from WM1361 cells and PBMCs were immunoprecipitated with anti-NKp44L mAb or the NKp44-Ig fusion protein, and then either (B) immunoblotted with the NKp44-Ig fusion protein or (C) anti-NKp44L mAb (αNKp44L). (B-C) Immunoprecipitations with isotype control are shown in the left panels. (D) WM1361 cell lysates were immunoprecipitated with the anti-N-ter polyclonal MLL5 Ab and immunoblotted with anti-NKp44L mAb (αNKp44L) or the NKp44-Ig fusion protein. C1 and C2 are controls obtained without lysate and without anti-MLL5 Ab, respectively. (E) Cell extracts from biotinylated WM1361 cells or PBMC were immunoprecipitated with anti-NKp44L mAb (αNKp44L) (left panel) or the NKp44-Ig fusion protein (right panel), and then immunoblotted with horseradish peroxidase-streptavidin. (F) Control Immunoprecipitation (IP) after cell-surface biotinylation of WM1361 cells. Cell extracts were immunoprecipitated with anti-actin mAb (α−actin) and then either immunoblotted with horseradish peroxidase-streptavidin (lower panel) or anti-actin mAb (α−actin) (higher panel). Nonimmunoprecipitated whole extract (WE) was immunoblotted with anti-actin mAb (α−actin). (C) Control obtained without lysate. IP, immunoprecipitation; WB, western blot. An arrow indicates the specific band.
NKp44L immune-detection. (A) Direct western blot in the presence of nonreduced or reduced lysates. Proteins from Jurkat cells were extracted with radioimmunoprecipitation assay (RIPA) or NET buffer, treated or untreated with DTT, resolved on an 8% SDS-PAGE and then immunoblotted with anti-NKp44L mAb (αNKp44L). (B-C) Cell lysates from WM1361 cells and PBMCs were immunoprecipitated with anti-NKp44L mAb or the NKp44-Ig fusion protein, and then either (B) immunoblotted with the NKp44-Ig fusion protein or (C) anti-NKp44L mAb (αNKp44L). (B-C) Immunoprecipitations with isotype control are shown in the left panels. (D) WM1361 cell lysates were immunoprecipitated with the anti-N-ter polyclonal MLL5 Ab and immunoblotted with anti-NKp44L mAb (αNKp44L) or the NKp44-Ig fusion protein. C1 and C2 are controls obtained without lysate and without anti-MLL5 Ab, respectively. (E) Cell extracts from biotinylated WM1361 cells or PBMC were immunoprecipitated with anti-NKp44L mAb (αNKp44L) (left panel) or the NKp44-Ig fusion protein (right panel), and then immunoblotted with horseradish peroxidase-streptavidin. (F) Control Immunoprecipitation (IP) after cell-surface biotinylation of WM1361 cells. Cell extracts were immunoprecipitated with anti-actin mAb (α−actin) and then either immunoblotted with horseradish peroxidase-streptavidin (lower panel) or anti-actin mAb (α−actin) (higher panel). Nonimmunoprecipitated whole extract (WE) was immunoblotted with anti-actin mAb (α−actin). (C) Control obtained without lysate. IP, immunoprecipitation; WB, western blot. An arrow indicates the specific band.
Expression of NKp44L by tumor and transformed cells
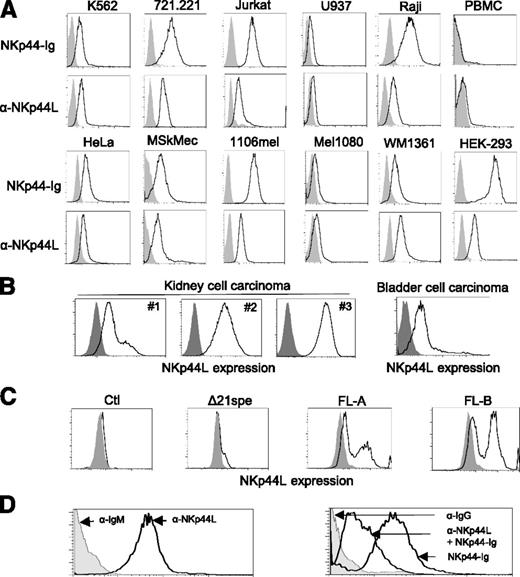
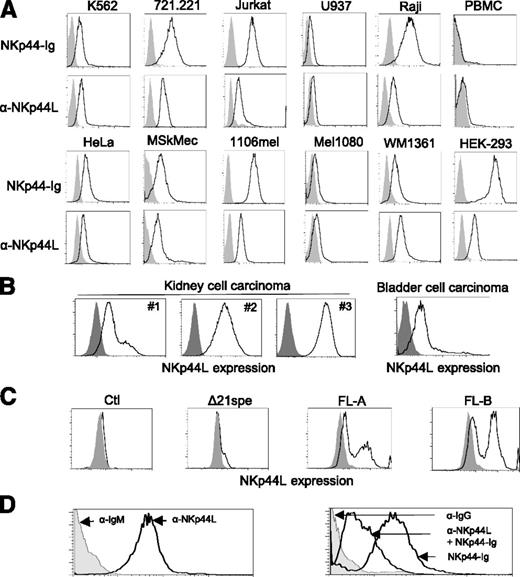
Next, we used flow cytometry to assess cell-surface expression of NKp44L in more than 40 different hematopoietic and nonhematopoietic cell lines, tested with either anti-NKp44L mAb or the NKp44-Ig fusion protein. NKp44L was specifically expressed in such hematologic transformed or tumor cell lines, as K562 (erythroleukemia), 721.221 (B-lymphoma), Jurkat, and MT2 (T-cell leukemia) cells, as well as in nonhematologic lines, such as HeLa (cervical carcinoma), MSKMec (endothelial), 1106mel, and WM1361 (melanoma) cells (Figure 3A) (also data not shown). In contrast, this ligand was barely detected at the cell surface either on U937 (promonocytic leukemia) or Mel1080 (melanoma) cells, and was not detected at all on Mono Mac (monocytic leukemia), LB33mel-B1 (melanoma), YTS (NK tumor), primary NK cell lines, or on resting peripheral blood from healthy donors (Figure 3A; supplemental Figure 3A). Importantly, the surface expression of NKp44L was also confirmed on primary tumor cell suspensions, obtained from patients with kidney and bladder malignancies (supplemental Methods). NKp44L cell surface expression was observed in all tumors with a frequency range between 20% and 100% in renal and 16% in bladder cell carcinoma (Figure 3B). These data are consistent with the cell-surface localization of NKp44L, determined by western blot analysis after cell-surface biotinylation (Figure 2E). Of note, intracellular expression of NKp44L was detected in all tested cell lines (supplemental Figure 3B).
NKp44L is a cell-surface ligand for NKp44. (A) Cell-surface expression of NKp44L was assessed by flow cytometry. The indicated tumor cell lines or primary PBMC were stained either with anti-NKp44L mAb and the NKp44-Ig fusion protein (bold lines) or with the corresponding controls (gray histograms). Data are representative of at least 3 independent experiments. (B) Cell-surface expression of NKp44L on kidney (from 3 different patients: patients 1, 2, and 3) and bladder cell carcinoma from primary biopsies. Tumor single-cell suspensions were stained with anti-NKp44L (bold lines), or with control (gray histograms). (C) NKp44L expression on mouse EL4-transfected cells. EL4 cells were stably transfected with the full-length (FL) sequence, the C-terminal-deleted (Δ21spe) sequence of NKp44L, or a control vector (Ctl). Transfected cells were drug-selected and then tested by flow cytometry with anti-NKp44L mAb (bold lines), or control (gray histograms). Two independent cell populations (FL-A and FL-B) expressing the full-length sequence of NKp44L were tested. (D) Blocking experiment on WM1361 cells. (Left panel) Staining with the anti-NKp44L mAb (bold line) or its IgM isotype control (grey histogram). (Right panel) Staining with the NKp44-Ig fusion protein, alone or in the presence of anti-NKp44L mAb (bold lines). IgG isotype control staining (grey histogram), serves as control.
NKp44L is a cell-surface ligand for NKp44. (A) Cell-surface expression of NKp44L was assessed by flow cytometry. The indicated tumor cell lines or primary PBMC were stained either with anti-NKp44L mAb and the NKp44-Ig fusion protein (bold lines) or with the corresponding controls (gray histograms). Data are representative of at least 3 independent experiments. (B) Cell-surface expression of NKp44L on kidney (from 3 different patients: patients 1, 2, and 3) and bladder cell carcinoma from primary biopsies. Tumor single-cell suspensions were stained with anti-NKp44L (bold lines), or with control (gray histograms). (C) NKp44L expression on mouse EL4-transfected cells. EL4 cells were stably transfected with the full-length (FL) sequence, the C-terminal-deleted (Δ21spe) sequence of NKp44L, or a control vector (Ctl). Transfected cells were drug-selected and then tested by flow cytometry with anti-NKp44L mAb (bold lines), or control (gray histograms). Two independent cell populations (FL-A and FL-B) expressing the full-length sequence of NKp44L were tested. (D) Blocking experiment on WM1361 cells. (Left panel) Staining with the anti-NKp44L mAb (bold line) or its IgM isotype control (grey histogram). (Right panel) Staining with the NKp44-Ig fusion protein, alone or in the presence of anti-NKp44L mAb (bold lines). IgG isotype control staining (grey histogram), serves as control.
Next we constructed a mammalian expression plasmid for the NKp44L full length ORF, and stably transfected it into mouse EL4 cells, a cell line negative for staining with anti-NKp44L mAb and resistant to NK lysis.27 Importantly, Figure 3C shows that 2 independent cell populations transfected with the NKp44L full-length (FL) ORF (FLA and FLB) are positive after staining with anti-NKp44L mAb. In contrast, NKp44L expression was not detected with anti-NKp44L mAb, in cells transfected with the control vector alone. Notably, in the cell population transfected by a vector expressing the C-terminal deleted protein (Δ21spe), no expression of NKp44L was detected at the cell surface (Figure 3C). Of note, similar intracellular staining with anti-NKp44L mAb was detected in EL4-transfected cells expressing either the full-length ORF or the C-terminal deleted protein (supplemental Figure 4A). Similar data were obtained after staining with the NKp44-Ig fusion protein (supplemental Figure 4B).
To confirm whether NKp44L is indeed a cell-surface ligand for NKp44, we also conducted blocking experiments on various cell lines. Figure 3D shows that the surface staining of the WM1361 cells by the NKp44-Ig fusion protein was markedly reduced when the cells were pre-incubated with anti-NKp44L mAb. Similar results were observed in other cell lines of various origins, including HeLa, 1106mel, and HEK-293 cells (supplemental Figure 5).
Cellular location of NKp44L
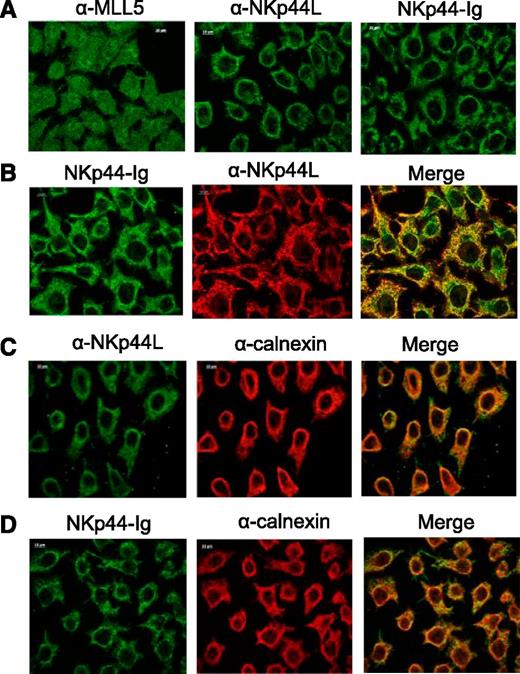
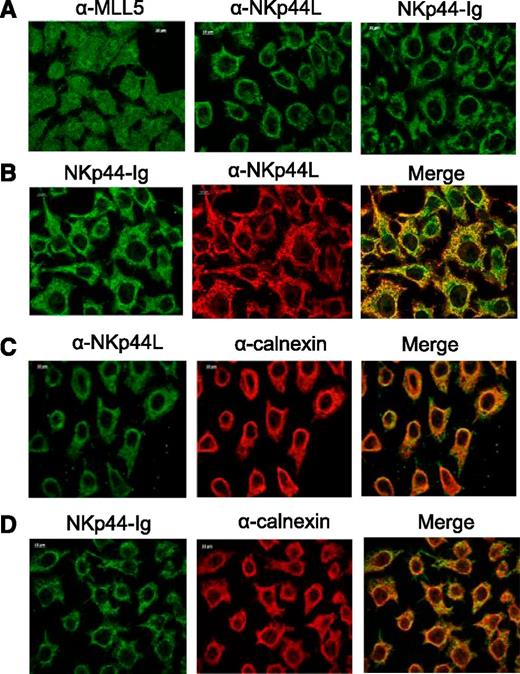
To reinforce these observations, we performed immunofluorescence staining in tumor cells to determine the subcellular localization of NKp44L. Because Deng et al28 previously reported that MLL5 is strictly nuclear, HeLa cells were fixed with acetone and stained with anti-MLL5 Ab, anti-NKp44L mAb, or the NKp44-Ig fusion protein. As expected, MLL5 was detected mainly in the nucleus, but also in the cytoplasm, whereas NKp44L was absent from the nucleus, but detected in the cytoplasm with both anti-NKp44L mAb and the NKp44-Ig fusion protein (Figure 4A; supplemental Figure 6). Similar results were obtained after fixation with ice-cold ethanol, and were reproduced with the melanoma cell line WM1361 (data not shown). As expected, most NKp44L staining colocalized with NKp44-Ig staining (Figure 4B; supplemental Figure 6), in agreement with the blocking experiments (Figure 3D). Importantly, a double-labeling with calnexin shows partial colocalization of NKp44L with the endoplasmic reticulum (Figure 4C), and a similar merge pattern was obtained with the NKp44-Ig fusion protein to detect NKp44L (Figure 4D; supplemental Figure 6). Notably, the subcellular localization of the Δ21spe protein in the EL4 transfectants revealed that this truncated form is also present in the nucleus, in contrast to the full-length form of NKp44L, which was excluded from it (supplemental Figure 4C).
Cellular localization of NKp44L in tumor cells. Immunofluorescence microscopy was performed on acetone-fixed HeLa cells stained with anti-MLL5 Ab (αMLL5), anti-NKp44L mAb (αNKp44L), or NKp44-Ig fusion protein. (A) Cytoplasmic and nuclear staining. (B) Intracellular colocalization of NKp44L and NKp44-Ig. (C-D) Intracellular colocalization of calnexin (α−calnexin) either with NKp44L (C) or NKp44-Ig fusion protein (D).
Cellular localization of NKp44L in tumor cells. Immunofluorescence microscopy was performed on acetone-fixed HeLa cells stained with anti-MLL5 Ab (αMLL5), anti-NKp44L mAb (αNKp44L), or NKp44-Ig fusion protein. (A) Cytoplasmic and nuclear staining. (B) Intracellular colocalization of NKp44L and NKp44-Ig. (C-D) Intracellular colocalization of calnexin (α−calnexin) either with NKp44L (C) or NKp44-Ig fusion protein (D).
NKp44L plays a functional role in cytotoxicity via the NKp44 receptor
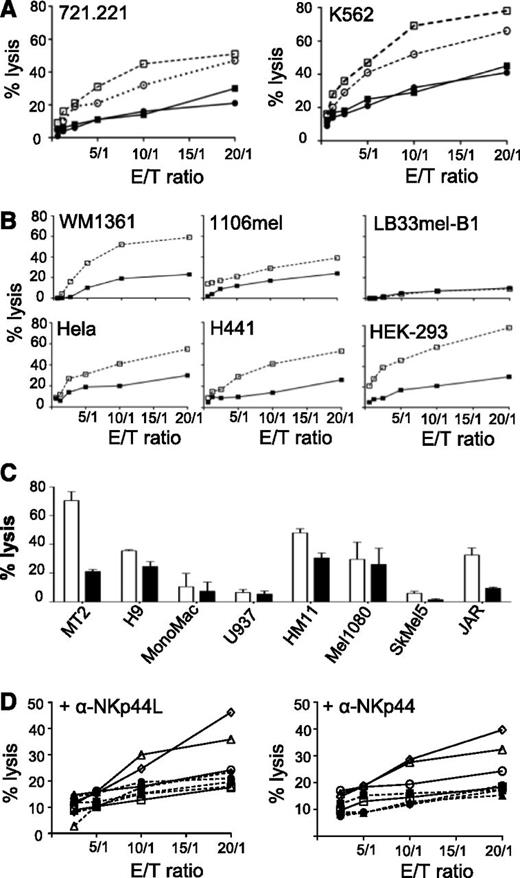
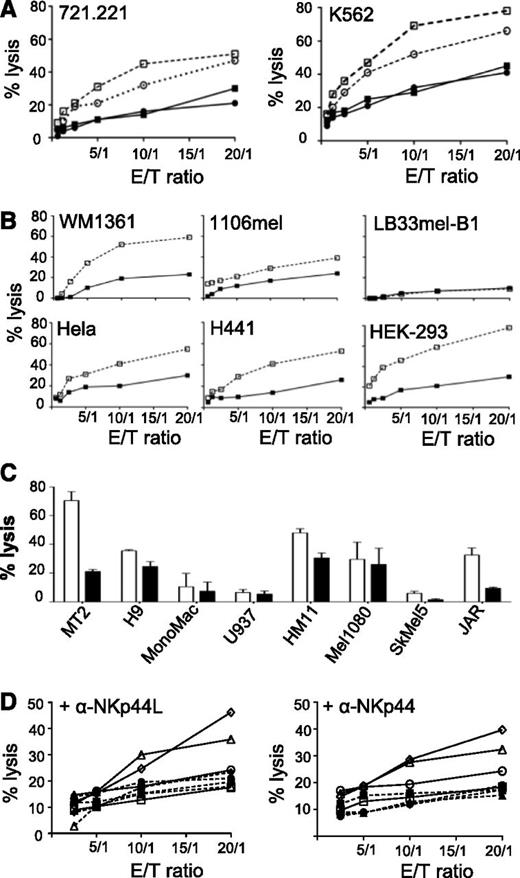
NKp44 is a triggering receptor involved in non-major histocompatibility complex (MHC)-restricted tumor lysis by activated NK cells.10,11 Purified NK cells from healthy donors were first activated by IL-2 treatment, and NKp44-induced cytotoxicity was then measured against different target cells. In the absence of anti-NKp44L mAb, NK cells effectively killed both classical MHC-class I negative 721.221 and K562 target cells. In the presence of anti-NKp44L mAb, lysis of these target cells was strongly inhibited (by as much as 40%) (Figure 5A). Similar inhibitory effects were also obtained after treatment of the effector NK cells with anti-NKp44 mAb (Figure 5A). We note that the inhibitory effect of anti-NKp44L mAb varied with the tumor cell line. Efficient inhibition (by more than 40%) was seen in some lines, including HEK293, WM1361, H441, and MT2 cells, and moderate inhibition (between 20% and 40%) in others, such as 1106mel, HeLa, and JAR cells. Finally, anti-NKp44L mAb did not significantly affect the killing of H9, U937, HM11, Mel1080, or SkMel5 cells (Figures 5B-C). Only a few tumor cell lines, none of which expressed NKp44L, were resistant to killing by IL2-activated NK cells; these included LB33mel-B1 and Mono Mac cells (Figures 5B-C). Such NK sensitivity was unrelated to the expression of class-I MHC molecules (data not shown), but was strongly linked to the cell-surface expression of NKp44L on the target cells (Figure 3).
Inhibition of natural cytotoxicity by anti-NKp44L mAb. Primary NK cells from healthy donors were purified, activated by IL-2 for 7 to 10 days, and then analyzed for cytotoxic activity against various tumors cell lines. (A) NK cytotoxicity of the 721.221 and K562 target cell lines, at different effector/target cell ratios. Open squares: IgM-isotype control-treated target cells. Closed squares: anti–NKp44L-treated target cells. Open circles: IgG1-isotype control-treated effector cells. Closed circles: anti-NKp44-treated effector cells. (B) Killing pattern of different target cells: WM1361, 1106mel, LB33mel-B1, HeLa, H441, and HEK-293 cell lines were tested for their sensitivity to NK lysis after treatment with anti-NKp44L mAb (closed squares) or IgM-isotype control (open squares), at different effector/target cell ratios. (C) NK lysis sensitivity of other hematologic (MT2, H9, Mono Mac, and U937), and nonhematologic (HM11, Mel1080, SkMel5, and JAR) tumor cells, after treatment with anti-NKp44L mAb (closed bars) or IgM-isotype control (open bars). Data are shown for an effector/target (E/T) ratio of 5:1. (D) Cytotoxicity of NK-resistant EL4 cells stably transfected with NKp44L. EL4 cells were either transfected either with the full length (triangles or diamonds), or the C-terminal-deleted (Δ21spe) (circles) sequence of NKp44L, or the control vector (squares). Transfected cells were tested for their NK sensitivity in the presence of IL2-activated NK cells without (plain lines and open symbols) or after pretreatment with specific mAbs (dotted lines and closed symbols). EL4 cells were treated (dotted lines) or not (plain lines) with anti-NKp44L mAbs (+αNKp44L) (left panel), whereas in the right panel, NK cells were treated (dotted lines) or not (plain lines) with anti-NKp44 mAbs (+αNKp44). More than 30% of CD3-CD56+ cells expressed NKp44. Data are representative of 2 independent experiments.
Inhibition of natural cytotoxicity by anti-NKp44L mAb. Primary NK cells from healthy donors were purified, activated by IL-2 for 7 to 10 days, and then analyzed for cytotoxic activity against various tumors cell lines. (A) NK cytotoxicity of the 721.221 and K562 target cell lines, at different effector/target cell ratios. Open squares: IgM-isotype control-treated target cells. Closed squares: anti–NKp44L-treated target cells. Open circles: IgG1-isotype control-treated effector cells. Closed circles: anti-NKp44-treated effector cells. (B) Killing pattern of different target cells: WM1361, 1106mel, LB33mel-B1, HeLa, H441, and HEK-293 cell lines were tested for their sensitivity to NK lysis after treatment with anti-NKp44L mAb (closed squares) or IgM-isotype control (open squares), at different effector/target cell ratios. (C) NK lysis sensitivity of other hematologic (MT2, H9, Mono Mac, and U937), and nonhematologic (HM11, Mel1080, SkMel5, and JAR) tumor cells, after treatment with anti-NKp44L mAb (closed bars) or IgM-isotype control (open bars). Data are shown for an effector/target (E/T) ratio of 5:1. (D) Cytotoxicity of NK-resistant EL4 cells stably transfected with NKp44L. EL4 cells were either transfected either with the full length (triangles or diamonds), or the C-terminal-deleted (Δ21spe) (circles) sequence of NKp44L, or the control vector (squares). Transfected cells were tested for their NK sensitivity in the presence of IL2-activated NK cells without (plain lines and open symbols) or after pretreatment with specific mAbs (dotted lines and closed symbols). EL4 cells were treated (dotted lines) or not (plain lines) with anti-NKp44L mAbs (+αNKp44L) (left panel), whereas in the right panel, NK cells were treated (dotted lines) or not (plain lines) with anti-NKp44 mAbs (+αNKp44). More than 30% of CD3-CD56+ cells expressed NKp44. Data are representative of 2 independent experiments.
Further proof that NKp44L is implicated in NK cytotoxicity came from sensitivity to lysis of NK-resistant mouse EL4 cells when stably transfected with NKp44L. These NKp44L-transfected EL4 cells were substantially more sensitive to NK lysis than nontransfected EL4 cells (supplemental Figure 4D). In contrast, EL4 cells transfected by a vector expressing the protein with its specific C-terminal motif deleted (Δ21spe) were sensitive as much as the cells transfected with the control vector (supplemental Figure 4D), consistently with their lack of NKp44L cell-surface expression after transfection (Figure 3C). Figure 5D (left panel) shows that treatment with anti-NKp44L mAbs almost entirely abolished the NK sensitivity of NKp44L-transfected EL4 cells. Similar inhibitory effects were obtained after treatment of the NK cells with anti-NKp44 mAb (Figure 5D, right panel). These data confirmed that NKp44L expression mediates sensitivity to NKp44+ NK cells.
Discussion
Here, we identified a cellular ligand for the activating NKp44 receptor, as a new shorter isoform of the MLL5 gene, which maps to chromosome 7q22 and is characterized by a specific C-terminal motif that interacts with NKp44 receptor. MLL5, also known as a lysine methyltransferase 2E is highly specific for lysine 4 of histone H3.29-31 MLL5 has been implicated in hematopoietic and myogenic differentiation.31-33 Overexpression and knockdown of MLL5 both induce cell cycle arrest at various phases, suggesting a versatile function of MLL5 throughout the cell cycle.28 In addition, its elevated expression was recently associated with favorable outcome in acute myeloid leukemia.34 To date, several other isoforms listed in supplemental Table 3 have been reported besides MLL5.35,36 Strikingly, compared with the other isoforms, NKp44L is characterized by a specific exon (21spe) at the C-terminus, probably implicated in its unique subcellular localization, as compared with the other MLL proteins. Importantly, the mRNA expression profiles of MLL5 and NKp44L are very different, pleotropic for MLL5, but significantly more restrictive to tumor cells for NKp44L.
We found that NKp44L encodes a highly conserved 1168-amino-acid protein. It is constitutively expressed on a broad panel of hematopoietic, and nonhematopoietic tumor and transformed cell lines of various origins, a finding supported by other groups using the soluble NKp44-Ig fusion protein.25 To date, however, we cannot rule out the possibility that other cellular ligands for NKp44 might be identified, as occurred for NKG2D.37 We previously reported that NKp44L is also expressed on bystander uninfected CD4+ T cells from HIV-infected patients.20-22,38 In these CD4+ T cells, the cell-surface translocation of NKp44L is mediated through a signaling cascade that involves sequential activation of PI3K, and NADPH oxidase, as well as TC10 inactivation.39 Saliently, MLL5 has been described as a nuclear protein,28 whereas NKp44L is not found in the nucleus, consistently with data of Madrid and Ganem.40 They also showed that during lytic Kaposi’s sarcoma-associated herpes virus infection, NKp44L is instead mislocalized and concentrated in the nucleus, whereas its expression on the cell surface decreases, as previously shown in chronic HIV infection.21,38
Intriguingly, no obvious transmembrane sequence or glycosylphosphatidylinositol anchor motif was deduced for the NKp44L sequence. Comparative experiments performed with the full-length and the C-terminal-deleted sequences of NKp44L strongly suggest that the C-terminal motif, implicated in the interaction with NKp44 receptor and located close to a putative glycosaminoglycan attachment site, could play a role in the cell-surface expression of NKp44L. We can hypothesis that NKp44L is associated to an unknown transmembrane coreceptor, or alternatively secreted into the extracellular matrix or retained at the cell surface trough sulfated glycosaminoglycan, as described for the proto-oncogene WNT141 or fibroblast growth factor-2.42 This last hypothesis could be consistent with previous data showing a direct binding of NKp44 with highly charged synthetic heparan sulfate,15 and with the three-dimensional structure of NKp44, showing that its inner groove surface is compatible to host an oligosaccharide.43 Thus, NKp44L exhibits a particular feature that points to nonconventional intracelllular trafficking that will require further analysis.
NKp44L presents characteristics notably similar to those observed with other NK activating receptor ligands, including MHC class-I related chain A/B and UL16 binding proteins, both of which are recognized by NKG2D,44 whereas B7-H6, a member of the B7 family protein, interacts with NKp30.18 NKp44L plays a role in cytotoxicity against various transformed and tumor cell lines, as demonstrated here by the sharp decrease of NK lysis activity in the presence of either anti-NKp44L or anti-NKp44 mAbs. The lytic effect is associated with the level of NKp44L expression on target cells, and might also depend on the density of receptors on the cell surface of different effector cells.10,11 Importantly, Gasser et al45 have described in detail several mechanisms, including major genotoxic stress that regulates expression of the NKG2D ligands. A parallel can be drawn with NKp44L, which is induced on normal CD4+ T cells after stress by H2O2 treatment.34 On the other hand, stimulation by cytokines (type I and II interferons, IL-2, IL-12, IL-15, IL-18, and transforming growth factor-β) does not affect the regulation of NKp44L (data not shown), but can induce some NKG2D ligands.46,47 These data suggest, similar to other ligands for activating NK receptors, that NKp44L transmits a danger signal in certain pathological situations, including certain infections and cancers, to induce the lysis of target cells by activated NKp44+ NK cells.
In conclusion, the identification of a cellular ligand for NKp44 highlights the role of self-molecules as a danger signal to alert the NK cell network. The near-total absence of NKp44L in normal tissues coupled with its expression in a broad array of pathological situations calls for prompt investigation of the regulation of its expression and could suggest a new approach for immunotherapeutic strategies.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof J. Strominger (Molecular and Cellular Biology Department, Harvard University, Cambridge, MA), Dr L.W. Deng (Department of Biochemistry, National University of Singapore, Singapore), Dr J. Boyson (Department of Medicine, University of Vermont, Burlington, VT), Dr O. Mandelboim (Hebrew University-Hadassah Medical School, Jerusalem, Israel), and Dr S. Sagan (Centre National de la Recherche Scientifique, Unité Mixte de Recherche 7203, Laboratoire des BioMolécules, Paris, France) for discussions and important suggestions, Prof M.O. Bitker (Service d’Urologie, Hôpital Pitié-Salpêtrière, Paris, France) for samples of renal and bladder carcinoma, and E. Perret of the Plate-Forme d’Imagerie Dynamique (Imagopole, Institut Pasteur, Paris, France) for technical assistance.
This study was supported in part by grants from the Institut National de la Santé et de la Recherche Médicale and the Université Pierre et Marie Curie (Paris-6) (V.V.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
V.V. is a researcher from the Centre National de la Recherche Scientifique, and the recipient of a Contrat Hospitalier de Recherche Translationnelle.
Authorship
Contribution: F.B., P.D., and V.V. conceived experiments; F.B., K.D., and V.V. designed experiments; F.B., A.S., K.D., and V.V. performed experiments; F.B., A.S., M.E., P.D., and V.V. analyzed the data; K.D. analyzed experiments; and V.V. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincent Vieillard, Department Immunity & Infections, INSERM UMR-S 945, Pitié-Salpêtrière Hospital, 83 Boulevard de l’Hôpital, Paris 75013, France; e-mail: vincent.vieillard@upmc.fr.