Abstract
Our group and others have reported the murine SHIP1-deficient mouse model of MDS/CMML is characterized by expansion of immunosuppressive, arginase 1 (Arg1)-positive M2-macrophages and myeloid-derived suppressor cells (MDSC) (Rauh et al., Immunity, 2005). Moreover, translational studies confirmed increased Arg1 in bone marrow aspirate cells of subsets of MDS and CMML patients (Rauh et al., Blood Abstract 2010: 1855), although involving impractical Arg1 enzymatic assays and Western blots. Herein, our goals were to 1) confirm this Arg1 immune signature in an independent cohort of patients, using more clinically applicable immunohistochemistry (IHC), 2) determine the associated risk profile with recent prognostic scoring systems, and 3) begin to connect this Arg1 immune signature with recurring MDS/CMML-acquired mutations.
With ethics approval, 29 BM biopsies (decalcified, FFPE) and clinical parameters were retrieved from the archives of Kingston General Hospital: 6 controls (4 negative lymphoma staging BM and 2 mild anemia NYD; mean age +/- std = 57 +/- 13 y), 13 MDS (1 RA, 1 RARS, 1 del(5q), 5 RCMD, 3 RAEB-1, 2 RAEB-2; 76 +/- 14 y), and 10 CMML patients (7 CMML-1, 3 CMML-2; 72 +/- 10 y). H&E and anti-human Arg1 IHC (1/2500 dilution, clone HPA003595, Sigma) were conducted under optimized, automated conditions (Ventana). IHC scoring was recorded independently by 2 blinded Hematopathologists. IPSS, IPSS-R (Greenberg), WPSS (Malcovati), CMML PSS (Such) and Mayo CMML (Patnaik) scores were calculated. Floxed TET2 and Vav-Cre mice were obtained from JAX and used according to approved Queen's University Animal Care protocols. TET2 sequences were obtained from genomic DNA using custom AmpliSeq primer pools and the Ion Torrent PGM platform (LifeTech). Linear regression and student's t-tests were conducted with Prism software (GraphPad).
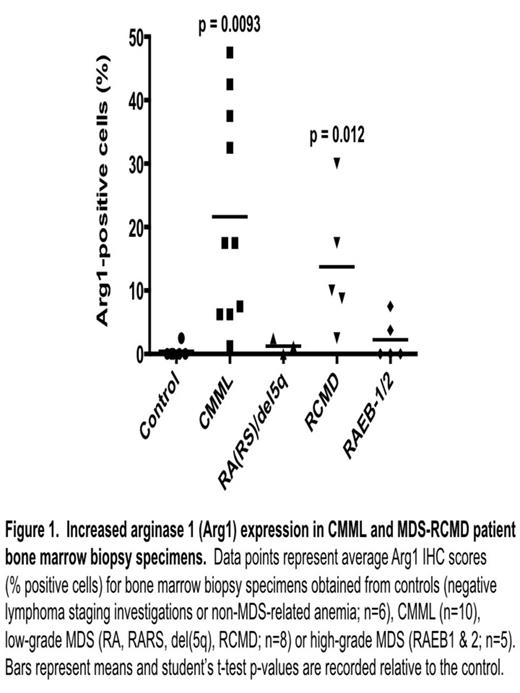
1) We demonstrated increased BM biopsy Arg1 IHC expression in CMML (22 +/- 17% Arg1-positive cells; n = 10; p = 0.0093) and low-grade MDS, particularly RCMD (14 +/- 11%; n = 5; p = 0.012) relative to control subjects (0.4 +/- 1%; n = 6) (Figure 1). Significantly increased mean Arg1 expression was not seen in other MDS subtypes (n = 8). These findings were consistent with our previously reported 40-subject (Toronto) BM aspirate Arg1 assay/Western blot cohort (Rauh et al. Blood Abstract 2010:1855), suggesting the reproducibility of these findings and the clinical utility of IHC-based assessment. Parallel Arg1 IHC is underway on the Toronto cohort, to determine correlations with enzymatic assays/Westerns.
2) We previously reported Arg1 over-expression in the Toronto cohort was significantly associated with the lowest IPSS and WPSS MDS clinical risk categories and now extend this to IPSS-R. In our Kingston cohort, only trends to lower MDS risk were observed. In contrast, increased Arg1 expression was not associated with clinical risk (CPSS and Mayo scores) in either CMML cohort. Thus, Arg1 expression associated with neutral risk in CMML and neutral to lower risk in MDS patients. Both the clinical risk profiles and proportions of Arg1 over-expressing MDS/CMML patients were reminiscent of reported TET2 mutation risk profiles/percentages.
3)TET2-deficient mice demonstrated increased monocytes, macrophages and CD11b+Gr1+ splenocytes (immuno-phenotypically consistent with MDSC), phenocopying SHIP1-deficient mice. We are currently determining whether TET2-/- mouse macrophages are similarly M2-skewed. In parallel, were are obtaining TET2 genomic sequences (along with other recurring mutated genes) for our Toronto and Kingston cohorts, in order to determine if the Arg1 immune signature associates with a particular mutation(s). These findings will be discussed.
1) Using two independent patient cohorts, we demonstrated significant Arg1 over-expression in CMML and low-grade (RCMD) MDS. Arg1 IHC warrants further investigation as an ancillary diagnostic test. 2) Increased Arg1 expression had neutral prognostic significance in CMML and neutral to low-risk MDS associations. 3) Increased Arg1 expression in MDS/CMML may be driven by mutant TET2, impacting the epigenetics of MDSC and M2-macrophage expansion, controlled by SHIP1 and related signaling networks. Confirmatory studies are in progress.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.