Abstract
The conversion of plasminogen (PLG) to plasmin is a critical step in the activation of the fibrinolytic system. Due to its broad substrate specificity, plasmin plays a pleotropic role in the circulation and extravascular space, controlling the degradation of fibrin clots as well as potential functions in regulating cell migration and angiogenesis. PLG activation is regulated through the interaction of tPA or uPA with plasminogen activator inhibitor type one (PAI-1). Genome-wide association studies (GWAS) have identified polymorphisms associated with the plasma levels of tPA and PAI-1, but to date, no studies have directly examined the control of PLG levels.
In order to identify genetic determinants of PLG levels we performed GWAS in a cohort of 3244 young healthy individuals (557 sibships) who participated in the Genes and Blood Clotting Study (GABC) or the Trinity Student Study (TSS).(Desch KC et al. PNAS 2013, 110:588-593) All subjects provided written informed consent. Details of the genotyping and data cleaning process for these cohorts are as previously described. (Desch KC et al. PNAS 2013, 110:588-593) PLG levels were measured in both cohorts by AlphaLISA (Perkin-Elmer, Waltham, MA). Levels were natural log transformed and adjusted for participant age, gender and BMI (TSS only) prior to use in heritability estimates and GWAS, which were performed using the PLINK, EMMAX, MERLIN, GCTA and METAL software packages.
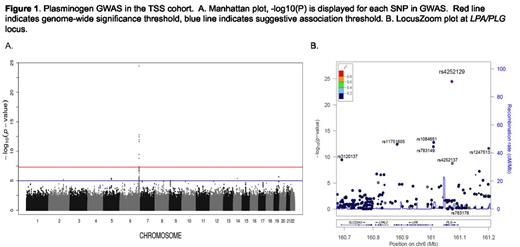
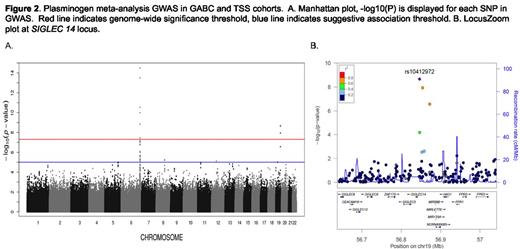
The median PLG levels were 101.7 IU/dL and 104.7 IU/dL in the GABC and TSS cohorts respectively and had a 1.7 fold variation between the 5th and 95th percentiles. The narrow-sense heritability using intra-class correlation was 59.7% based on 1277 siblings in GABC and TSS, which was similar to the estimate of 60.2% based on the genotype data and the MERLIN software package. GWAS for PLG in the European only subset of GABC (n=940) did not reveal any genome-wide significant signals. However, GWAS for PLG levels in TSS (n=2304) revealed 9 significant SNPs on chromosome 6q26 at the PLG (plasminogen) and LPA (lipoprotein A) locus that together explained 7.8% of the variance in PLG levels according to the GCTA method, Figure 1. The top SNP was rs4252129 (p = 3.47E-25) which results in a single amino acid substitution (R523W) in PLG. The next strongest associations were with two SNPs near LPA, rs1084651 and rs783149 (p = 1.60E-13 and 1.07E-12 respectively). These SNPs are located between LPA and PLG (Figure 1). To determine if the LPA SNPs were independent from the top PLG SNP, a conditional analysis was performed using rs4252129 as a covariate. Rs1084651 and rs783149 remained significant (p= 7.73E-16 and 6.48E-15 respectively), verifying that they represent a second, independent association signal. Meta-analysis of both cohorts (using 723,715 common SNPs) confirmed the 6q26 signal and revealed another significantly associated region on 19q13.41, Figure 2. Two SNPs, rs10412972 and rs11084102 (p = 2.16E-09 and 1.14E-08, respectively) are intergenic variants 4.8 and 12.6 Kb downstream from SIGLEC14. The significant SNPs in the meta-analysis, 6 on 6q26 and 2 on 19q13.41, explained 5.8% of the variance for PLG. The top SNP in the TSS GWAS, rs4252129, had a minor allele frequency (MAF) of 1.4% in GABC and was not included in the GABC GWAS or meta-analysis.
Ginsburg:Shire plc: Membership on an entity’s Board of Directors or advisory committees; Portola Pharmaeuticals: Membership on an entity’s Board of Directors or advisory committees; Catalyst Biosciences: Membership on an entity’s Board of Directors or advisory committees; Merck pharmaceuticals: Consultancy; Boston Children's Hospital: From Baxter to Children's Hosp (VWF), From Baxter to Children's Hosp (VWF) Patents & Royalties; University of Michigan: From Baxter to University of Michigan (ADAMTS13) Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.