Abstract
Acute Myeloid Leukemia (AML) is a form of cancer that affects the cells of the hematopoietic system. The currently applied WHO classification of genetically defined AML subtypes based on cytogenetic and molecular aberrations of known oncogenes and tumor suppressors allows to stratify patients into genetically defined low-, intermediate, and high-risk groups. Yet, this detailed genetic information has rarely resulted in the development of targeted therapies that are directed at the underlying genetic lesions, or even personalized therapies. With the notable exceptions of chronic myeloid leukemia (CML) and acute promyelocytic leukemia (APL), the prognosis for most AML subtypes treated with standard chemotherapy regimens remains for the most part poor with a median overall survival of approximately 18 months.
The development of cancer may have both genetic and epigenetic causes, but what is common to all cancers is a unique gene-expression pattern (GEP) that drives the malignant phenotype and thus partly differs from that of normal cells representing the same type of cells/tissue and developmental stage. Significantly the unique GEP of cancer cells have been illustrated in a multitude of gene-expression profiling studies where mRNA were quantified using microarray or deep sequencing platforms. These technologies have been outstanding to group, diagnose and forecast AML subtypes based on differences in GEPs. However, they have only demonstrated a marginal role in the identification of potential therapeutic targets. This is mainly due to the fact that these studies have compared AMLs with other AMLs and not with corresponding normal cells of the same developmental stages. This is an obvious prerequisite if one wants to define the genes that drives the malignant phenotype of AML cells.
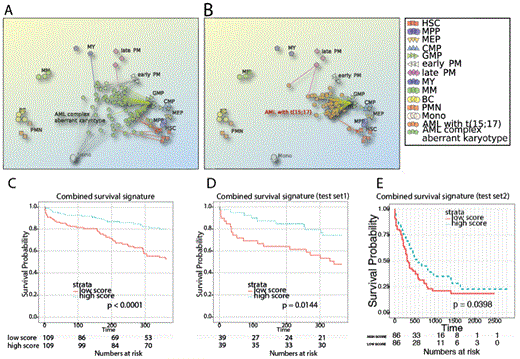
(A, B) PCA plots of individual GEP profiles from two AML subtypes mapped to the normal landscape of hematopoietic differentiation. (C-E) Stratification of three independent patient cohorts with survival gene signature generated by comparison of cancer and direct normal counterpart.
(A, B) PCA plots of individual GEP profiles from two AML subtypes mapped to the normal landscape of hematopoietic differentiation. (C-E) Stratification of three independent patient cohorts with survival gene signature generated by comparison of cancer and direct normal counterpart.
Our method resulted in improved clinical prognostication of AML patients and enabled us to identify new potential therapeutic target genes and oncogenic pathways activity that are over-expressed in AML cells as compared to their normal counterparts. Similarly, using the connectivity-map database (a collection of genome-wide transcriptional expression data from cultured human cells treated with bioactive small molecules), we could screen substances that could reverse the aberrant gene-expression signatures; some of these drugs are already in use in AML treatment or in clinical trials, which strengthens the validity of our bioinformatics method. A singularly interesting perspective in this regard is the possibility to tailor individualized drug combinations potentially targeting cancer cells synergistically by combined inhibition of multiple oncogenic targets and pathways.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.