In this issue of Blood, Papaemmanuil et al present results from the largest single targeted sequencing effort in myelodysplastic syndrome (MDS) to date both in terms of number of patients studied (738 patients, of which 603 had MDS) and genes sequenced (111 genes).1
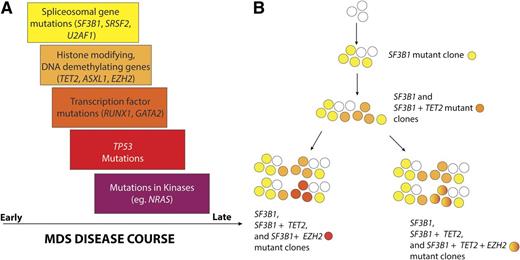
Temporal order of mutation acquisition in MDS based on data from Papaemmanuil et al. (A) Analysis of the variant allele fractions in 111 genes sequenced in 738 patients with MDS and related disorders yielded a common pattern of mutation acquisition in the natural history of MDS. Based on this work, it appears that mutations in genes encoding spliceosomal components occur early in the pathogenesis of MDS with mutations in epigenetic modifiers and transcripitional regulators occurring later. Mutations activating cytokine signaling appear to be among the latest clonal events in MDS. In the figure, the width of the box indicates the time frame in which mutations of genes described in the box occurs. The overlap of the boxes indicates the fact the patterns of mutation acquisition in MDS are not absolute. (B) The authors noted that nearly half of MDS patients had ≥2 frequently mutated genes simultaneously. In these individuals, one-third had multiple subclones and an attempt to construct the clonal hierarchy of 1 such individual bearing TET2, SF3B1, and EZH2 mutations from the study is shown.
Temporal order of mutation acquisition in MDS based on data from Papaemmanuil et al. (A) Analysis of the variant allele fractions in 111 genes sequenced in 738 patients with MDS and related disorders yielded a common pattern of mutation acquisition in the natural history of MDS. Based on this work, it appears that mutations in genes encoding spliceosomal components occur early in the pathogenesis of MDS with mutations in epigenetic modifiers and transcripitional regulators occurring later. Mutations activating cytokine signaling appear to be among the latest clonal events in MDS. In the figure, the width of the box indicates the time frame in which mutations of genes described in the box occurs. The overlap of the boxes indicates the fact the patterns of mutation acquisition in MDS are not absolute. (B) The authors noted that nearly half of MDS patients had ≥2 frequently mutated genes simultaneously. In these individuals, one-third had multiple subclones and an attempt to construct the clonal hierarchy of 1 such individual bearing TET2, SF3B1, and EZH2 mutations from the study is shown.
Recently, comprehensive mutational profiling has identified the importance of somatic mutations in predicting outcome2,3 and characterizing clonal hierchical events in patients with MDS.4-6 Data from Papemmanuil et al confirm and extend prior observations about the correlation of mutations with clinical phenotype in MDS and present a clearer picture of the stepwise transformation events most commonly seen in MDS (see figure).
Given the relatively recent discovery of many recurrently mutated genes in MDS and the need for validation of prior mutational correlative data, a number of clinically important observations are now solidified with the publication of this work. First, it is clear that the total number of mutations present in a patient with MDS has important prognostic value, even independent of currently clinically used prognostic information.1,2 Second, of the most commonly mutated genes in MDS, mutations in ASXL1, SRSF2, RUNX1, and TP53 are once again associated with worsened outcome and decreased leukemia-free survival.2,3 Comprehensively sequencing such a large cohort of patients also further strengthens our knowledge of the mutational combinations that are likely critical in MDS pathogenesis. For instance, by carefully the examining mutational co-occurrences, Papaemmanuil et al have identified that each of the mutated spliceosomal genes has a unique spectrum of mutational overlap. This might account for the divergent phenotypes associated with each of the spliceosomal mutations despite their mutual exclusivity,3,7 a hypothesis that will be excellent to test functionally.
In addition to these observations, Papaemmanuil et al also examined the mutational allele frequency of each of the commonly mutated genes to estimate the temporal order of mutation acquisition in MDS and understand the impact of such subclonal mutations (defined as mutations not present in the entire proportion of malignant cells) on disease course. The authors found that the detection of mutations in genes associated with adverse outcome had significant impact even when not present in the major clone. This finding simplifies the clinical interpretation of poor prognostic mutations and emphasizes the need for sensitive detection methods to identify them in practice. This is reminiscent of the recently appreciated importance of subclonal genetic events in chronic lymphocytic leukemia (CLL),8 another leukemia characterized by a highly diverse clinical course and a frequent clinical practice of monitoring disease without initiation of therapy. Unlike this recent work in CLL, or prior work in MDS,5,6 which used sequencing of all coding exomes or the entire genome to track cancer clones, Papaemmanuil et al used targeted sequencing of the most frequently mutated genes at a much higher depth to achieve a similar goal. These data suggest that mutations impacting RNA splicing and DNA methylation occur early in disease progression with mutations affecting histone post-translational modifications and DNA demethylation following, and kinase activating mutations (such as KIT and NRAS) occurring even later in disease progression.4 Indeed, we previously identified that mutations in NRAS can be present at surprisingly low mutation allele burdens (0.02-0.1%) and are associated with adverse overall survival even when present at 0.5% mutant allele burden.9
Despite the large number of patients studied and extent of sequencing performed, it is clear that there is still much to be learned about clonal dynamics and molecular prognostication in MDS. For example, surprisingly, 22% of the 738 patients included here had no identifiable somatic genetic abnormality (mutation or cytogenetic alteration). Secondly, prior work using whole genome sequencing (WGS) to define the clonal architecture and events in the progression of MDS to acute myeloid leukemia identified the presence of dozens to hundreds of mutations in a founding clone followed by outgrowth of ≥1 subclone each marked by the acquisition of an increasing number of new mutations.5,6 Comparing this WGS approach with a targeted sequencing approach, it appeared that using variant allele frequencies of recurrently mutated genes alone did not fully recapitulate clonal architecture identified by WGS of the same individuals.6 This may account for why the observations of mutational acquisition suggested from the data here are not absolute. Further work should be done via both longitudinal sequencing studies as well as functional studies to understand the potential importance, if any, on the order of acquisition of commonly occurring mutations in MDS.
The authors compared the predictive value of clinical prognostic measures, such as those considered by the International Prognostic Scoring System (IPSS), to the presence of mutations. In their global analysis, they found that the addition of mutation information did not significantly refine the predictive power of a model considering clinical variables alone, although it did improve on the IPSS. More careful analysis, potentially distinguishing different molecular subtypes of MDS, will be needed to determine how best to incorporate mutational information into the prediction of prognosis.
Finally, this study describe how extensive targeted sequencing provides reliable copy number data that completely coincides with clinical fluorescence in situ hybridization(FISH)/cytogenetic data in MDS. From a purely practical standpoint, this suggests that serial sequencing might simultaneously provide both mutational data and copy number data capable of supplanting recurrent FISH/cytogenetic testing in MDS where balanced translocations are rare.
Based on the above, it is very likely that longitudinal genetic characterization of MDS will add to our ability to predict clinical outcomes, as well as identify emerging clones with potentially actionable alterations. To fully take advantage of what we learn by sequencing, we will have to pair genetic results to specific treatments, something that will require more therapeutic options for patients with MDS than we have today. In the meantime, we can apply this data toward making better prognostic models and improving our understanding of biological processes that underlie these often morbid disorders. Further work to identify the effect of existing treatments, such as DNA hypomethylating agents and allogeneic transplantation, on the genetic characteristics of persistent and relapsed disease clones, is likely to be incredibly informative and of great value to patients.
Conflict-of-interest disclosure: The authors declare no competing financial interests.