Key Points
Programmed death ligands 1 and 2 are rearranged at a frequency of 20% in PMBCL.
Abstract
The pathogenesis of primary mediastinal large B-cell lymphoma (PMBCL) is incompletely understood. Recently, specific genotypic and phenotypic features have been linked to tumor cell immune escape mechanisms in PMBCL. We studied 571 B-cell lymphomas with a focus on PMBCL. Using fluorescence in situ hybridization here, we report that the programmed death ligand (PDL) locus (9p24.1) is frequently and specifically rearranged in PMBCL (20%) as compared with diffuse large B-cell lymphoma, follicular lymphoma, and Hodgkin lymphoma. Rearrangement was significantly correlated with overexpression of PDL transcripts. Utilizing high-throughput sequencing techniques, we characterized novel translocations and chimeric fusion transcripts involving PDLs at base-pair resolution. Our data suggest that recurrent genomic rearrangement events underlie an immune privilege phenotype in a subset of B-cell lymphomas.
Introduction
Primary mediastinal large B-cell lymphoma (PMBCL) is an aggressive disease known to share certain genotypic and phenotypic features with classic Hodgkin lymphoma (CHL) and diffuse large B-cell lymphoma (DLBCL).1,2 However, the complete landscape of genetic alterations involved in PMBCL pathogenesis has yet to be fully elucidated.3 Among the most common chromosomal alterations in PMBCL are amplifications of chromosome 9p and translocations involving CIITA (16p13.13).4-7 These aberrations have been suggested to affect tumor-microenvironment interactions resulting in immune privilege.7,8 Here, we demonstrate that rearrangements involving immune cell anergy-inducing programmed death ligand (PDL) 1 (CD274) and 2 (PDCD1LG2) are recurrent in and characteristic of PMBCL. Furthermore, we show such rearrangements are correlated with elevated transcript levels, and we characterize novel translocations identified using high-throughput sequencing.
Study design
We studied 571 primary B-cell lymphoma samples in conjunction with 17 established B-cell-derived cell lines. Using in-house bacterial artificial chromosome probes, fluorescence in situ hybridization (FISH) or FISH combined with CD30 immunofluorescence (in the case of CHL specimens) was performed to characterize the PDL locus.7,9,10 These cases were also analyzed with Epstein-Barr virus (EBV)-encoded RNA in situ hybridization. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed for CD274 and PDCD1LG2 transcript expression on a subset of cases from the FISH cohort (N = 76). Surface PDCD1LG2 expression of the cell lines was determined via flow cytometry. To characterize the Hodgkin lymphoma cell lines found to be rearranged by FISH, 1 whole-transcriptome sequencing (RNA-seq) library (CHL-derived L-428) was reanalyzed, and 1 new whole-genome library (L-428) and 2 new RNA-seq libraries (CHL-derived L-1236 and nodular lymphocyte predominant Hodgkin lymphoma-derived DEV) were sequenced.7 This study was approved by the BC Cancer Agency (REB: H11-00684) and conducted in accordance with the Declaration of Helsinki.
Results and discussion
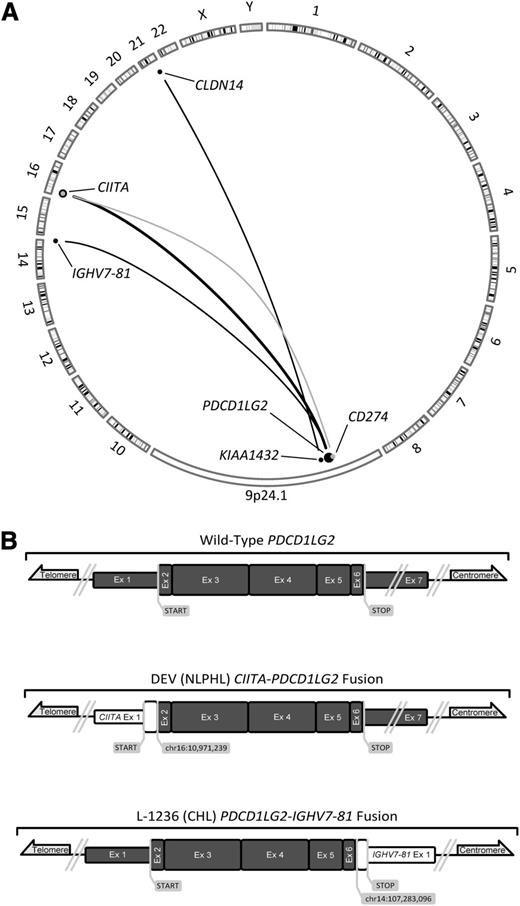
There is increasing evidence that genes involved in tumor-microenvironment interactions play a crucial role in the pathogenesis of B-cell lymphomas.8 Two such genes, CD274 and PDCD1LG2, have been implicated in promoting tumor cell immune evasion.11,12 These ligands have been reported to be overexpressed in solid tumors, including lymphomas, but the mechanisms by which overexpression occurs are incompletely understood.4,7,13-15 Using an in-house break-apart FISH assay (supplemental Figure 1; supplemental Table 1), we determined the frequency of PDL locus aberration in 17 lymphoma-derived cell lines (supplemental Table 2) and 571 clinical lymphoma samples (Figure 1). This analysis revealed a break-apart frequency of 20% and an amplification frequency of 29% in 125 PMBCL cases. The prevalence of PMBCL break-apart events was significantly higher than in DLBCL, PCNSL, TDLBCL, and FL (P < .05). No significant differences of any noted clinical parameters, including treatment outcome variables (progression-free and overall survival), were observed between PDL rearranged, amplified, and nonrearranged cases.
Frequency of PDL locus alterations and correlation with transcript expression levels. (A) Representative FISH signal patterns from 2 clinical samples; top: break-apart (primary PMBCL), bottom: amplification (primary PMBCL). (B) The proportion of PDL locus (9p24.1) aberration across 7 different subtypes of B-cell lymphomas observed using an in-house break-apart FISH assay (N values: DLBCL = 134; PCNSL = 130; primary testicular DLBCL [TDLBCL] = 82; PMBCL = 125; CHL = 20; nodular lymphocyte predominant Hodgkin lymphoma [NLPHL] = 12; follicular lymphoma [FL] = 68). P values were < .05 for the number of break-apart cases in PMBCL in comparison with lymphomas with N > 12; P < .05 for the number of amplified cases in PMBCL in comparison with lymphomas with N > 20. (C) CD274 transcript expression in 17 cell lines and 76 clinical samples as determined via qRT-PCR (N values: PMBCL = 48; DLBCL = 19; TDLBCL = 6; PCNSL = 3). The dotted red line represents basal CD274 expression as determined by reactive tonsil cells. The break-apart to neutral comparison reached statistical significance (P = .03). Amplified to neutral (P = .001) and amplified to gain (P = .01) comparisons also reached statistical significance. (D) PDCD1LG2 transcript expression in the cohort described above. The dotted blue line represents basal PDCD1LG2 expression. Statistical significance was researched between break-apart to neutral (P = .0003), break-apart to gain (P = .001), and break-apart to amplified (P = .005) case comparisons. The amplified to neutral comparison also reached statistical significance (P = .002). (E) Differential expression of both PDL transcripts in break-apart cases. Dots circled in red (2) were Sanger-validated to have a translocation involving the CD274 locus, whereas those in blue (5) were found to harbor rearrangements of PDCD1LG2. Note how break-apart events result in disparate expression levels between transcripts.
Frequency of PDL locus alterations and correlation with transcript expression levels. (A) Representative FISH signal patterns from 2 clinical samples; top: break-apart (primary PMBCL), bottom: amplification (primary PMBCL). (B) The proportion of PDL locus (9p24.1) aberration across 7 different subtypes of B-cell lymphomas observed using an in-house break-apart FISH assay (N values: DLBCL = 134; PCNSL = 130; primary testicular DLBCL [TDLBCL] = 82; PMBCL = 125; CHL = 20; nodular lymphocyte predominant Hodgkin lymphoma [NLPHL] = 12; follicular lymphoma [FL] = 68). P values were < .05 for the number of break-apart cases in PMBCL in comparison with lymphomas with N > 12; P < .05 for the number of amplified cases in PMBCL in comparison with lymphomas with N > 20. (C) CD274 transcript expression in 17 cell lines and 76 clinical samples as determined via qRT-PCR (N values: PMBCL = 48; DLBCL = 19; TDLBCL = 6; PCNSL = 3). The dotted red line represents basal CD274 expression as determined by reactive tonsil cells. The break-apart to neutral comparison reached statistical significance (P = .03). Amplified to neutral (P = .001) and amplified to gain (P = .01) comparisons also reached statistical significance. (D) PDCD1LG2 transcript expression in the cohort described above. The dotted blue line represents basal PDCD1LG2 expression. Statistical significance was researched between break-apart to neutral (P = .0003), break-apart to gain (P = .001), and break-apart to amplified (P = .005) case comparisons. The amplified to neutral comparison also reached statistical significance (P = .002). (E) Differential expression of both PDL transcripts in break-apart cases. Dots circled in red (2) were Sanger-validated to have a translocation involving the CD274 locus, whereas those in blue (5) were found to harbor rearrangements of PDCD1LG2. Note how break-apart events result in disparate expression levels between transcripts.
To study the correlation of genomic rearrangements and copy-number changes of the PDL locus with gene expression, we performed qRT-PCR on 17 cell lines and 76 clinical cases that had fresh frozen material available (supplemental Table 3). These cases were stratified according to the genomic aberration status as determined by FISH. In break-apart positive cases across all lymphomas, PDCD1LG2 transcript levels were significantly higher as compared with neutral (P = .0003), gained (P = .001), and amplified (P = .005) loci (Figure 1; supplemental Table 4). A significant difference was also observed between CD274 transcript levels of rearranged and copy-neutral cases (P = .03), although only a trend was found when compared with copy-number aberrated cases. This reduced level of correlation may be attributable to the heterogeneity in rearrangement anatomy, which disparately affects the PDL loci. The observation that CD274 and PDCD1LG2 expression levels were also elevated (relative to PDL expression in reactive tonsil cells) in cases where no rearrangement could be observed via FISH, suggests alternative mechanisms of deregulation exist. These mechanisms might include epigenetic and microRNA regulatory factors.16,17 As EBV infection has been implicated in PDL expression, we performed EBV-encoded RNA in situ hybridization (supplemental Table 1).18 No significant correlation was observed between EBV positivity and either PDL–break-apart or transcript levels, although the small number of EBV-positive cases in our study limits statistical power.
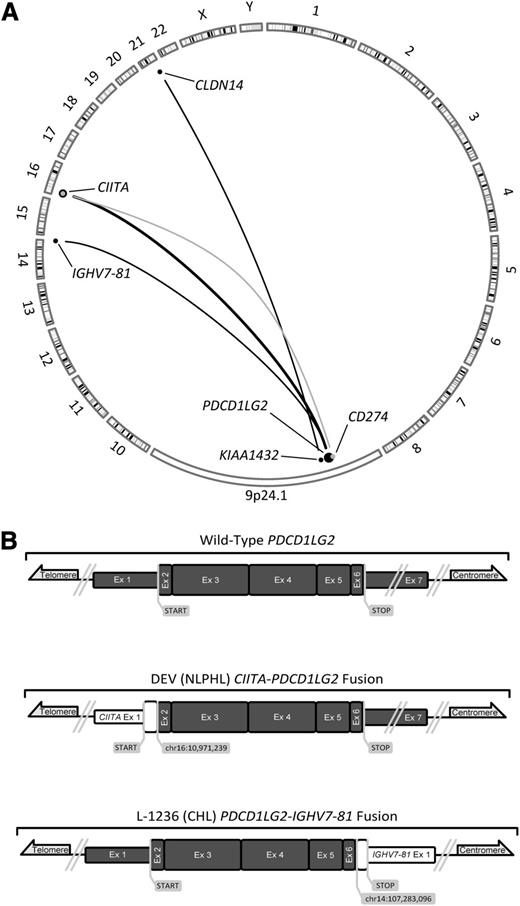
To determine the rearrangement structures that were observed at the PDL locus, we reanalyzed previously published RNA-seq data of L-428.7 Additionally, we sequenced the whole genome of L-428 and the whole transcriptomes of L-1236 and DEV. Using genomic breakpoint and fusion transcript predicting algorithms, we mined these data with a specific focus on the 9p24.1 locus (supplemental Tables 5 and 6). We identified 3 novel fusion transcripts within this locus that were validated by Sanger sequencing: CIITA-PDCD1LG2 (DEV), KIAA1432-CLDN14 (L-428), and PDCD1LG2-IGHV7-81 (L-1236) (Figure 2). The fusion transcript observed in DEV was found to be similar to 2 fusions that we had previously reported in clinical PMBCL cases.7 Although clearly involving the 9p24.1 locus, the FISH break-apart signal pattern of L-428 was attributed to a translocation 60 kb downstream of PDCD1LG2. The mechanism by which this translocation increases PDL expression is unknown; although KIAA1432-JAK2 fusions have been recently implicated in small cell lung cancer.19 The chimeric transcript produced by L-1236 was predicted to generate a C-terminal-truncated PDCD1LG2 protein. In this instance, the receptor binding site, coded in the fourth exon, was left intact.20
A summary of known translocations involving the 9p24.1 locus in PMBCL and 2 novel fusion transcripts. (A) Translocations involving CD274 are depicted in gray, whereas those that are involved or situated downstream from PDCD1LG2 are depicted in black. Dot size and adjoining line thickness qualitatively depict the number of cases involving those loci; the CIITA-PDCD1LG2 line represents 4 cases. Three of the 6 depicted translocations are novel, PDCD1LG2-IGHV7-81, CIITA-PDCD1LG2, and KIAA1432-CLDN14. (B) Sanger-validated fusion transcripts involving PDCD1LG2; all 7 exons of PDCD1LG2 are depicted in dark gray and are in scale to one another. Known and suspected protein coding regions are drawn with a greater exon width.
A summary of known translocations involving the 9p24.1 locus in PMBCL and 2 novel fusion transcripts. (A) Translocations involving CD274 are depicted in gray, whereas those that are involved or situated downstream from PDCD1LG2 are depicted in black. Dot size and adjoining line thickness qualitatively depict the number of cases involving those loci; the CIITA-PDCD1LG2 line represents 4 cases. Three of the 6 depicted translocations are novel, PDCD1LG2-IGHV7-81, CIITA-PDCD1LG2, and KIAA1432-CLDN14. (B) Sanger-validated fusion transcripts involving PDCD1LG2; all 7 exons of PDCD1LG2 are depicted in dark gray and are in scale to one another. Known and suspected protein coding regions are drawn with a greater exon width.
To investigate the effect of these rearrangements on protein expression, we performed flow cytometry on several cell lines with a specific focus on those found harboring 9p24.1 translocations: L-428, L-1236, and DEV. We found elevated expression of PDCD1LG2 in all 3 rearranged cell lines compared with U-HO1 and DOHH-2 (normal loci) (supplemental Figure 2). Furthermore, expression levels in L-428 and L-1236 were determined to be in agreement with the literature.4 PDCD1LG2 protein surface levels were highest in DEV and were also appreciably greater than in cell line MEDB-1 (amplified locus). This shows that a PDCD1LG2 epitope is readily detectable in DEV cells at elevated levels corresponding to a CIITA-PDCD1LG2 fusion that may alter T-cell activity states in the microenvironment, as previously demonstrated.7
By merging the data of this study with the 3 PDL fusions previously reported by our group (Figure 2), we have validated the presence of 5′ fusion transcripts involving PDL in immortalized cells, and we have established the existence of a novel fusion transcript that involves the 3′ end of PDCD1LG2.7 As substantially elevated transcript levels were observed in all instances with validated direct involvement of PDL rearrangements, upregulated expression of PDLs may be a consistent mechanism that contributes to pathogenesis. Deregulated transcript expression is believed to arise via juxtaposing either a highly active promoter (CIITA) or enhancing elements (IGHV7-81) adjacent to PDLs.7,21,22 Other potential selective advantages that these translocations confer may be related to the altered function of the involved fusion partner genes. This would include decreased tumor immunogenicity as a result of reduced major histocompatibility complex II expression from CIITA-associated translocations.7 Another potential benefit specific to the 3′ translocation in L-1236 is the loss of the transcript 3′ untranslated region. This may lead to a loss of microRNA binding sites increasing the half-life of PDL transcript and/or the loss of the exons coding for the transmembrane domain region, which may act to solubilize the protein.17,23,24
Taken together, our findings establish that rearrangement of the PDL locus is recurrently selected for and that such rearrangements lead to elevated transcript levels and/or the production of chimeric PDL fusion transcripts. Recently, phase 1/2 clinical trials targeting the PDL pathway in solid tumors have shown promising outcomes.14,15,25 Our data strongly suggest that PDLs and their high-affinity receptor, programmed death 1 (PDCD1), are rational drug targets in a subgroup of B-cell lymphomas characterized by PDL locus aberrations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (111043) (R.D.G.) and the Terry Fox Research Institute (Terry Fox New Frontiers Program in Cancer [1023]) (C.S.), and generous support from the British Columbia Cancer Foundation.
C.S. and S.P.S. are recipients of Career Investigator Awards from the Michael Smith Foundation for Health Research; A.M. is supported by a Mildred-Scheel-Cancer-Foundation Fellowship; and D.D.W.T. is supported by UBC fellowships and the Canadian Hematology Society.
Authorship
Contribution: D.D.W.T. designed and performed research, and interpreted data; F.C.C., R.S.L., and A.W.M. analyzed sequencing data and contributed to the figures; S.B.-N., B.W.W., K.L.T., A.M., and G.W.S. produced and analyzed data; J.G., R.K., and A.T. assisted with data collection; D.W.S., K.J.S., and S.P.S. provided editorial input; R.D.G. analyzed data, constructed the database, and edited the paper; C.S. designed research, analyzed data, and approved the paper; and D.D.W.T. and C.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Steidl, Department of Experimental Therapeutics, BC Cancer Research Centre, 675 West 10th Ave, Vancouver, BC, Canada, V5Z 1L3; e-mail: csteidl@bccancer.bc.ca.

![Figure 1. Frequency of PDL locus alterations and correlation with transcript expression levels. (A) Representative FISH signal patterns from 2 clinical samples; top: break-apart (primary PMBCL), bottom: amplification (primary PMBCL). (B) The proportion of PDL locus (9p24.1) aberration across 7 different subtypes of B-cell lymphomas observed using an in-house break-apart FISH assay (N values: DLBCL = 134; PCNSL = 130; primary testicular DLBCL [TDLBCL] = 82; PMBCL = 125; CHL = 20; nodular lymphocyte predominant Hodgkin lymphoma [NLPHL] = 12; follicular lymphoma [FL] = 68). P values were < .05 for the number of break-apart cases in PMBCL in comparison with lymphomas with N > 12; P < .05 for the number of amplified cases in PMBCL in comparison with lymphomas with N > 20. (C) CD274 transcript expression in 17 cell lines and 76 clinical samples as determined via qRT-PCR (N values: PMBCL = 48; DLBCL = 19; TDLBCL = 6; PCNSL = 3). The dotted red line represents basal CD274 expression as determined by reactive tonsil cells. The break-apart to neutral comparison reached statistical significance (P = .03). Amplified to neutral (P = .001) and amplified to gain (P = .01) comparisons also reached statistical significance. (D) PDCD1LG2 transcript expression in the cohort described above. The dotted blue line represents basal PDCD1LG2 expression. Statistical significance was researched between break-apart to neutral (P = .0003), break-apart to gain (P = .001), and break-apart to amplified (P = .005) case comparisons. The amplified to neutral comparison also reached statistical significance (P = .002). (E) Differential expression of both PDL transcripts in break-apart cases. Dots circled in red (2) were Sanger-validated to have a translocation involving the CD274 locus, whereas those in blue (5) were found to harbor rearrangements of PDCD1LG2. Note how break-apart events result in disparate expression levels between transcripts.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/13/10.1182_blood-2013-10-535443/4/m_2062f1.jpeg?Expires=1765935658&Signature=d~awJy42JuWdkScsBJ1YxWZP6oK7Gwdlfw8xNlphtEtZRcK7LZFypSDluMsoHI93NWbwhP~6TW8da90~D3McJR0NJvdMrT2KeBxJUMX4b44plLvnn2H4U0NsqoMLWzjbeiwCjyHxxKMtzU32REY4V1LsKq8VkqsCtXMQGNwjm~M1BfYnGjGO43bWWgrBhVjgP69oYm3YZyxg5t9tU8wr42OhW4SQaH5Bf373-V3L62a0vpzs38e5nVjtymy0x6v0sba9eOEYuFnNSgqMy1VnEmgxLuMDsPa2D7vtPceMbT~LAO4kUhsmJHD3Z6HyEGGaCV-4eud3kcC647RFzFoH~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. Frequency of PDL locus alterations and correlation with transcript expression levels. (A) Representative FISH signal patterns from 2 clinical samples; top: break-apart (primary PMBCL), bottom: amplification (primary PMBCL). (B) The proportion of PDL locus (9p24.1) aberration across 7 different subtypes of B-cell lymphomas observed using an in-house break-apart FISH assay (N values: DLBCL = 134; PCNSL = 130; primary testicular DLBCL [TDLBCL] = 82; PMBCL = 125; CHL = 20; nodular lymphocyte predominant Hodgkin lymphoma [NLPHL] = 12; follicular lymphoma [FL] = 68). P values were < .05 for the number of break-apart cases in PMBCL in comparison with lymphomas with N > 12; P < .05 for the number of amplified cases in PMBCL in comparison with lymphomas with N > 20. (C) CD274 transcript expression in 17 cell lines and 76 clinical samples as determined via qRT-PCR (N values: PMBCL = 48; DLBCL = 19; TDLBCL = 6; PCNSL = 3). The dotted red line represents basal CD274 expression as determined by reactive tonsil cells. The break-apart to neutral comparison reached statistical significance (P = .03). Amplified to neutral (P = .001) and amplified to gain (P = .01) comparisons also reached statistical significance. (D) PDCD1LG2 transcript expression in the cohort described above. The dotted blue line represents basal PDCD1LG2 expression. Statistical significance was researched between break-apart to neutral (P = .0003), break-apart to gain (P = .001), and break-apart to amplified (P = .005) case comparisons. The amplified to neutral comparison also reached statistical significance (P = .002). (E) Differential expression of both PDL transcripts in break-apart cases. Dots circled in red (2) were Sanger-validated to have a translocation involving the CD274 locus, whereas those in blue (5) were found to harbor rearrangements of PDCD1LG2. Note how break-apart events result in disparate expression levels between transcripts.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/13/10.1182_blood-2013-10-535443/4/m_2062f1.jpeg?Expires=1765935659&Signature=rEqE4mRAAhUdI5mEtibDzBpl7eLenw8VdWC-3kCeC1hnPlGuneXEIOWkapujRQc3ozIKW5ZjoPnIZHzdF2HG8IW6r6CyEywqJejaMN8aqVv382RG7tdIDwtXDzaNiljuZ--k9Gt~nT720~y3xHqSIce9sruGdogxTfaYTCQSVQUgDzfuv8rPrsVJSENSxdhDF~JOxTm7qd9nXWXnO-NHLESlukr~AWf34lvM1thuXLA6w6LeW7zmKTaeCmJXZS7nak~wZ61fecaKSC8xeCxuOeV7q08psctt6epGUl0tbvXdjXzFcdB978HaIu2X-uquptE~6ZVqyYfNSf3MRdChJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)