Key Points
Hyperactivated STAT5a binds LEF-1 protein leading to NLK/NARF/ubiquitin-dependent degradation of LEF-1 followed by defective granulopoiesis.
In CN patients, elevated levels of phospho-STAT5a resulted in diminished LEF-1 expression, which could be restored by bortezomib treatment.
Abstract
The transcription factor lymphoid enhancer–binding factor 1 (LEF-1), which plays a definitive role in granulocyte colony-stimulating factor (G-CSF) receptor-triggered granulopoiesis, is downregulated in granulocytic progenitors of severe congenital neutropenia (CN) patients. However, the exact mechanism of LEF-1 downregulation is unclear. CN patients are responsive to therapeutically high doses of G-CSF and are at increased risk of developing acute myeloid leukemia. The normal expression of LEF-1 in monocytes and lymphocytes, whose differentiation is unaffected in CN, suggests the presence of a granulopoiesis-specific mechanism downstream of G-CSF receptor signaling that leads to LEF-1 downregulation. Signal transducer and activator of transcription 5 (STAT5) is activated by G-CSF and is hyperactivated in acute myeloid leukemia. Here, we investigated the effects of activated STAT5 on LEF-1 expression and functions in hematopoietic progenitor cells. We demonstrated that constitutively active STAT5a (caSTAT5a) inhibited LEF-1–dependent autoregulation of the LEF-1 gene promoter by binding to the LEF-1 protein, recruiting Nemo-like kinase and the E3 ubiquitin-ligase NARF to LEF-1, leading to LEF-1 ubiquitination and a reduction in LEF-1 protein levels. The proteasome inhibitor bortezomib reversed the defective G-CSF–triggered granulocytic differentiation of CD34+ cells from CN patients in vitro, an effect that was accompanied by restoration of LEF-1 protein levels and LEF-1 messenger RNA autoregulation. Taken together, our data define a novel mechanism of LEF-1 downregulation in CN patients via enhanced ubiquitination and degradation of LEF-1 protein by hyperactivated STAT5.
Introduction
Granulocyte colony-stimulating factor (G-CSF) regulates the survival, proliferation, and maturation of granulocytic precursors.1 The G-CSF receptor (G-CSFR) is a member of the type I cytokine receptor family that triggers the phosphorylation of receptor-associated protein tyrosine kinases, including Janus kinase 1/2 (JAK1/2) and members of the Src kinase family (eg, Lyn, Syk).2,3 The phosphorylation of these tyrosine kinases leads to activation of a cascade of downstream effector molecules, such as signal transducer and activator of transcription (STAT) proteins, or causes recruitment of various adaptor proteins. These, in turn, mediate activation of downstream pathways, including phosphoinositide 3-kinase/Akt, Ras/Raf/mitogen-activated protein kinase (MAPK, and nicotinamide phosphoribosyltransferase/NAD+/sirtuin 1.4-6
Among the different STATs, STAT3 and STAT5 are robustly activated by G-CSFR, but in a different manner and serving different effector functions. STAT3 is activated in a sustained fashion, but activation of STAT5 is transient, with maximal activation levels occurring within 10 to 30 minutes.7-10 STAT5 is involved in the maintenance and expansion of human hematopoietic stem/progenitor cells and is crucial for cell survival and proliferation.11 In myeloid progenitors lacking both STAT5a and STAT5b, the clonal advantage conferred by mutant G-CSFR was found to be abrogated.12 Stress-induced erythropoiesis is severely impaired in STAT5−/− mice.13 In line with this, constitutive activation of STAT5 results in impaired in vitro myelopoiesis of human hematopoietic stem/progenitor cells in association with downregulation of myeloid-associated factors such as C/EBPα.14,15 Moreover, hyperactivation of STAT5 signaling has been implicated in various hematological malignancies, including B-cell receptor-ABL–induced chronic myeloid leukemia and acute myeloid leukemia (AML), and in myeloproliferative disorders, such as chronic myelomonocytic leukemia and polycythemia vera.9,16
STAT5 augments or inhibits the promoter activity of target genes by either direct DNA binding of dimeric or tetrameric forms or through cooperation with other cofactors bound nearby.9 However, several studies have highlighted a potential role of STAT5 in DNA binding–independent transcriptional repression via protein–protein interactions or through competition for or sequestration of limited cofactors.17 Thus, STAT5 mediates repression of activator protein 1 independent of DNA binding.18 Moreover, activation of STAT5 through tyrosine phosphorylation is required for the inhibition of glucocorticoid receptor–dependent transcription through glucocorticoid receptor/STAT5 complex formation. Importantly, recent studies from liver endothelial cells of STAT5−/− mice have indicated a role for STAT5 in negatively regulating granulopoiesis through direct or indirect repression of G-CSF expression.19 Moreover, impaired hematopoietic differentiation in acute promyelocytic leukemia has been linked to an altered pattern of co-repressor recruitment to retinoic acid receptor α and its dissociation by stimulation with all-trans retinoic acid; the latter effect is mediated by the STAT5 moiety of the STAT5-retinoic acid receptor α fusion protein that is expressed in this condition.20
Severe congenital neutropenia (CN) is characterized by a maturation arrest of granulopoiesis at the promyelocytic stage. G-CSFR signaling is severely impaired in CN patients. CN patients show a response to G-CSF therapy; however, pharmacologically high doses of G-CSF (1-100 μg/kg body weight per day) are required to increase neutrophil counts.21 CN patients are at increased risk of developing AML (cumulative incidence, ∼15-21%).22 Whether and how G-CSF affects this predisposition remains unclear. Long-term therapy with pharmacological doses of G-CSF or elevated levels of endogenous G-CSF in plasma of CN patients23 may cause genomic instability because of increased pressure on cell division and DNA replication. Moreover, G-CSF may lead to a preferential secondary outgrowth of preexisting cell clones containing mutations, for example in the G-CSFR (CSF3R) gene.24 Hematopoietic cells from nearly 80% of CN patients who developed leukemia show truncations in the cytoplasmic region of the G-CSFR crucial for maturation signaling.25,26
We have previously shown that the transcription factor lymphoid enhancer–binding factor 1 (LEF-1) plays a definitive role in G-CSF–triggered granulopoiesis.27 LEF-1 and its target genes, including the myeloid-specific transcription factor C/EBPα, are severely downregulated in promyelocytes of CN patients. However, the mechanism underlying LEF-1 downregulation has remained elusive. LEF-1 belongs to the LEF-1/T-cell factor (TCF) family of high-mobility group domain–containing transcription factors. Members of the LEF-1/TCF transcription factor family generally act through the canonical Wnt signaling pathway in a functional complex with β-catenin. Wnt-/β-catenin–independent stimuli, such as transforming growth factor-β and Notch pathways, may also engage LEF-1.28,29 Among the proteins known to interact with LEF-1 are β-catenin, ALY, paired-like homeodomain transcription factor 2, and Groucho/transducin-like enhancer of split.27-29 Interestingly, Nemo-like kinase (NLK) was shown to antagonize Wnt/β-catenin signaling by interaction with LEF-1 and LEF-1 phosphorylation.30,31 Recent studies have demonstrated that NLK-associated ring finger protein (NARF) interacts with LEF-1, leading to LEF-1 ubiquitination and degradation via the proteasome pathway.32 Specifically, it was shown that NARF complexes with NLK under steady-state conditions and, in response to an unknown ligand, acts through activation of NLK to exert E3 ubiquitin-ligase activity toward LEF-1.32 Interestingly, Kojima et al demonstrated that NLK is activated by G-CSF and interleukin (IL)-6 via STAT3.33 It has been shown that NLK interacts with STAT3 and that STAT3 activates NLK, serving as a scaffold between activated IL-6 and NLK.33,34 Whether hyperactivated STAT5 plays a similar scaffolding role in G-CSF–triggered NLK activation and NLK/NARF-mediated ubiquitination of LEF-1 is unclear.
Ubiquitination has emerged as an important modification that couples proteasome activity to chromatin remodeling and transcriptional regulation.35-38 Bortezomib, an inhibitor of the 26S proteasome, is a well-known therapeutic agent used clinically in combination with cell-cycle inhibitors to treat several leukemias, including AML.39 Here, we investigated the effects of activated STAT5 on LEF-1 expression and functions in hematopoietic progenitor cells. We further tested the effects of bortezomib on LEF-1 expression and defective G-CSF–triggered granulocytic differentiation of CD34+ cells from CN patients in vitro.
Materials and methods
Patients and control subjects
Nine patients participated in this study: 3 CN patients harboring HAX1 mutations, 3 CN patients with ELA2 mutations, and 3 patients with ELANE (n = 2) and HAX1 (n = 1) mutations who developed CN/AML. All subjects had received long-term (>1 year) G-CSF treatment; G-CSF doses ranged from 2.5 to 7.5 μg/kg per body weight daily, or once every 2 days. Three healthy volunteers received G-CSF at a dose of 5 μg/kg per body weight per day for 3 days. Bone marrow samples were collected in association with the annual follow-up recommended by the Severe Chronic Neutropenia International Registry. This study was approved by the Institutional Review Board of Hannover Medical School, and informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
Cell purification and separation
Bone marrow CD34+ cells were obtained by sequential immunomagnetic labeling of bone marrow mononuclear cells with anti-CD34+ MACS beads (Miltenyi Biotech) followed by positive selection. The purity of sorted CD34+ cells was greater than 96% as assessed by fluorescence-activated cell sorting (FACS) analysis.
Cell stimulation, intracellular protein staining, and FACS analysis
For the analysis of phospho-STAT5 levels, CD34+ cells were isolated from the bone marrow of CN patients and healthy individuals and used for analysis. For G-CSF stimulation, cells were incubated in ex vivo medium without serum and cytokines; after 2 hours, cells were treated with 10 ng/mL of recombinant human G-CSF for 10 minutes, washed in ice-cold phosphate-buffered saline/1% bovine serum albumin, fixed in 4% paraformaldehyde for 10 minutes, permeabilized with 0.5% Triton X-100 for 10 minutes, incubated with Alexa Fluor 647–conjugated anti-phospho (Y694)-STAT5 (BD Phosflow; BD Biosciences) antibodies for 1 hour at 4°C and analyzed by flow cytometry. Surface G-CSFR expression was analyzed by FACS using anti-human CD114 antibody (catalog number 346108) from Biolegend.
Western blot analysis
The following primary antibodies were used: rabbit polyclonal anti-STAT5a (Santa Cruz Biotechnology), rabbit monoclonal anti-phospho (Y694)-STAT5 (Cell Signaling Technology), rabbit monoclonal anti-LEF-1 (Cell Signaling Technology), mouse monoclonal anti-LEF-1 (CalBiochem), mouse monoclonal anti-β-actin (Santa Cruz Biotechnology), and rabbit polyclonal anti-α-tubulin (Cell Signaling Technology). Horseradish peroxidase–conjugated anti-mouse or anti-rabbit secondary antibodies (Santa Cruz Biotechnology) was used as appropriate.
Transfection of HEK293T cells
HEK293T cells were transfected with pcDNA 3.1-LEF1 alone or in combination with pcDNA 3.1 zeocin-caSTAT5a, or WT STAT5a or STAT5a Y-F MUT expression plasmids using Lipofectamine 2000 (Invitrogen), according to manufacturer’s protocol. After 36 hours, cells were treated or not with bortezomib alone or in combination with cycloheximide at the indicated concentrations and incubated for an additional 12 hours. dimethylsulfoxide, added at the same final concentration as present in treated samples was used as a vehicle control. Whole-cell lysates were obtained by lysing a defined number of cells in Laemmle sample buffer. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10% gels and transferred to polyvinylidene difluoride membranes. Blots were probed by incubating with primary antibodies overnight at 4°C.
Statistical analysis
Differences in mean values between groups were analyzed by 2-sided, unpaired Student t test using the SPSS V. 9.0 statistical package.
Results
Elevated G-CSF–triggered STAT5 phosphorylation in myeloid cells of CN patients
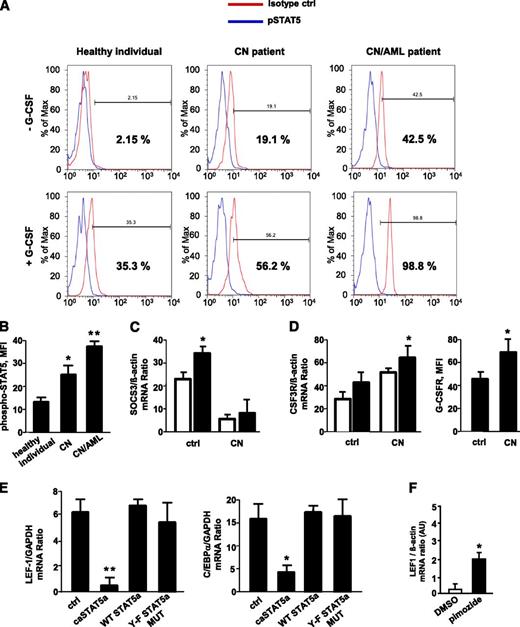
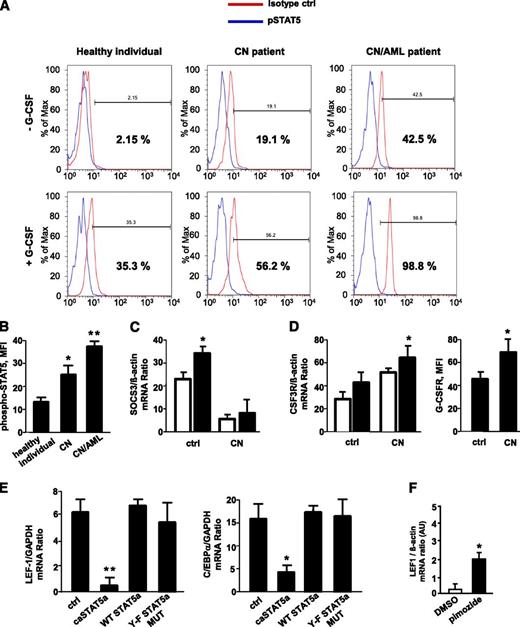
CD34+ cells from healthy individuals, CN patients harboring ELA2 (n = 3) or HAX1 (HCLS1-associated protein X-1; n = 3) mutations, and 3 CN patients with ELANE (n = 2) and HAX1 (n = 1) mutations who developed AML were stimulated with G-CSF, and phospho-STAT5 protein levels were measured. The levels of phosphorylated (activated) STAT5 were significantly elevated in unstimulated CD34+ cells from CN patients compared with those in healthy individuals; the relative increase was even greater in CN/AML patients. Stimulation with G-CSF induces phosphorylation of STAT5 in healthy individuals, an effect that was markedly higher in CN and CN/AML patients (Figure 1A-B). Previously, we reported elevated levels of JAK2 in myeloid cells of CN patients.40 Here, we found diminished messenger RNA (mRNA) expression levels of suppressor of cytokine signaling 3 (SOCS3; Figure 1C) as well as elevated G-CSFR mRNA and protein expression (Figure 1D) in CN myeloid cells compared with healthy individuals, a result in line with elevated phospho-STAT5. This enhanced phosphorylation, in turn, would be predicted to lead to STAT5 hyperactivation.
G-CSF–triggered phosphorylation of STAT5 is elevated in myeloid cells of CN patients; caSTAT5a inhibits mRNA expression of LEF-1 and its target gene C/EBPα. The levels of phospho-STAT5 in myeloid cells were assessed by FACS analysis of CD34+ cells from healthy individuals (n = 4), CN patients harboring ELA2 (n = 3) or HAX1 (n = 3) mutations, and CN patients who developed AML (n = 3) after incubating with or without recombinant human G-CSF (10 ng/mL) for 10 minutes. Cells were immunostained with an anti-phospho-STAT5 (Y694) antibody and isotype control antibody. (A) Representative histograms showing phSTAT5 (blue) and isotype (red) staining are depicted. (B) Bars show mean fluorescence intensity (MFI) of phospho-STAT5 (Y694) staining in CD34+ cells treated with G-CSF as described previously. Data represent means ± standard deviation (SD) and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (C) SOCS3 mRNA expression in CD33+ cells from G-CSF–treated healthy individuals (n = 3) and G-CSF–treated CN patients (n = 6) was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). SOCS3 mRNA levels were normalized to those of β-actin and are presented as arbitrary units (AUs). Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (D) G-CSFR mRNA and G-CSFR protein surface expression in CD33+ cells of G-CSF–treated healthy individuals (n = 3) and G-CSF–treated CN patients (n = 6). mRNA expression was measured by qRT-PCR. G-CSFR mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (E) CD34+ hematopoietic progenitor cells were transduced with a retroviral expression construct for caSTAT5a, WT STAT5a, STAT5a Y-F MUT, or ctrl-rv. After 60 hours of culture in X-VIVO 10 medium (Cambrex) supplemented with 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL thrombopoietin, 50 ng/mL stem cell factor, 50 ng/mL Flt3 ligand, and 10 ng/mL G-CSF, green fluorescent protein–positive cells were sorted, and LEF-1 and C/EBPα mRNA levels were measured by qRT-PCR. LEF-1 and C/EBPα mRNA levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase and are expressed as AUs. G-CSFR surface expression was measured by FACS. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (F) CD34+ cells from 1 CN patient were incubated with 5 μM pimozide, a specific STAT5 inhibitor, for 48 hours, and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). DMSO, dimethylsulfoxide.
G-CSF–triggered phosphorylation of STAT5 is elevated in myeloid cells of CN patients; caSTAT5a inhibits mRNA expression of LEF-1 and its target gene C/EBPα. The levels of phospho-STAT5 in myeloid cells were assessed by FACS analysis of CD34+ cells from healthy individuals (n = 4), CN patients harboring ELA2 (n = 3) or HAX1 (n = 3) mutations, and CN patients who developed AML (n = 3) after incubating with or without recombinant human G-CSF (10 ng/mL) for 10 minutes. Cells were immunostained with an anti-phospho-STAT5 (Y694) antibody and isotype control antibody. (A) Representative histograms showing phSTAT5 (blue) and isotype (red) staining are depicted. (B) Bars show mean fluorescence intensity (MFI) of phospho-STAT5 (Y694) staining in CD34+ cells treated with G-CSF as described previously. Data represent means ± standard deviation (SD) and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (C) SOCS3 mRNA expression in CD33+ cells from G-CSF–treated healthy individuals (n = 3) and G-CSF–treated CN patients (n = 6) was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). SOCS3 mRNA levels were normalized to those of β-actin and are presented as arbitrary units (AUs). Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (D) G-CSFR mRNA and G-CSFR protein surface expression in CD33+ cells of G-CSF–treated healthy individuals (n = 3) and G-CSF–treated CN patients (n = 6). mRNA expression was measured by qRT-PCR. G-CSFR mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (E) CD34+ hematopoietic progenitor cells were transduced with a retroviral expression construct for caSTAT5a, WT STAT5a, STAT5a Y-F MUT, or ctrl-rv. After 60 hours of culture in X-VIVO 10 medium (Cambrex) supplemented with 20 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL thrombopoietin, 50 ng/mL stem cell factor, 50 ng/mL Flt3 ligand, and 10 ng/mL G-CSF, green fluorescent protein–positive cells were sorted, and LEF-1 and C/EBPα mRNA levels were measured by qRT-PCR. LEF-1 and C/EBPα mRNA levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase and are expressed as AUs. G-CSFR surface expression was measured by FACS. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (F) CD34+ cells from 1 CN patient were incubated with 5 μM pimozide, a specific STAT5 inhibitor, for 48 hours, and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). DMSO, dimethylsulfoxide.
caSTAT5a inhibits LEF-1 and C/EBPα mRNA expression
The expression levels of LEF-1 and C/EBPα transcription factors are severely downregulated in myeloid cells of CN patients.27 LEF-1 expression was also undetectable in CD34+ cells from CN/AML patients studied here (data not shown). To test the role of STAT5a in this phenomenon, we measured LEF-1 and C/EBPα mRNA levels in CD34+ hematopoietic progenitor cells transduced with retroviral constructs expressing (1) WT STAT5a; (2) caSTAT5a, containing H299→R and S711/716→F mutations (STAT5a1*6) and possessing constitutive tyrosine kinase activity; (3) STAT5a Y-F MUT, in which tyrosine 694 of WT STAT5a was mutated; or (4) control retrovirus (ctrl-rv). We found a dramatic downregulation of LEF-1 and C/EBPα in caSTAT5a-transduced cells compared with samples transduced with WT STAT5a, Y-F STAT5a MUT, or ctrl-rv constructs (Figure 1E). Proliferation ratio and cellular composition were comparable between ctrl-rv– and caSTAT5a-rv–transduced groups (supplemental Table 1, available on the Blood Web site).
Moreover, incubation of CN hematopoietic cells in vitro with the specific STAT5 inhibitor, pimozide, restored LEF-1 mRNA expression, suggesting an important role of phosphorylated STAT5a in the downregulation of LEF-1 (Figure 1F).
caSTAT5a inhibits the transcriptional activity of the LEF-1 promoter
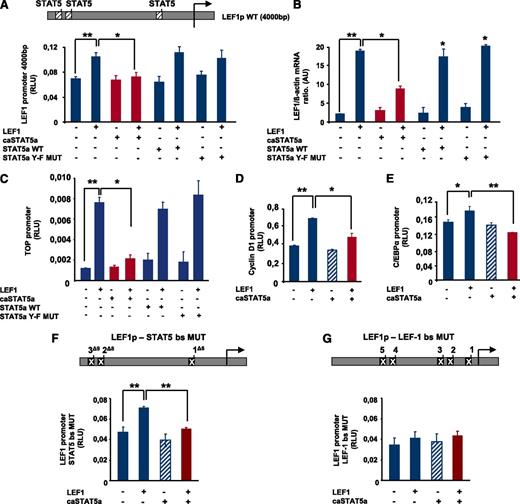
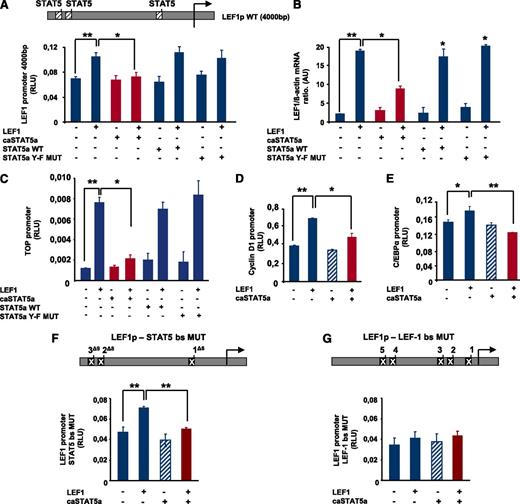
To analyze the effects of hyperactivated STAT5 on LEF-1 promoter activity, we generated a reporter gene construct comprising 4000 bp of the human LEF-1 promoter, which contains 3 STAT5 binding sites and 5 LEF-1 binding sites (Figure 2A). Co-transfection of HEK293 cells with a LEF-1 expression plasmid and the LEF-1 promoter-luciferase reporter construct resulted in upregulation of LEF-1 promoter activity, confirming LEF-1 autoregulation (Figure 2A). Co-transfection with caSTAT5a and LEF-1 expression plasmids inhibited LEF-1–dependent autoregulation of the LEF-1 promoter, whereas transfection with caSTAT5 alone had no effect (Figure 2A). Moreover, co-transfection with wild-type (WT) STAT5a or STAT5a Y-F MUT had no inhibitory effect on LEF1-mediated LEF-1 promoter autoregulation (Figure 2A). In line with this, caSTAT5a, but not WT STAT5a or STAT5a Y-F MUT, inhibited the LEF-1–mediated increase in LEF-1 mRNA levels (Figure 2B).
caSTAT5a inhibits the transcriptional activity of the LEF-1 promoter. (A) A LEF-1 reporter construct containing a 4000-bp upstream region of the LEF-1 gene harboring 3 binding sites identified in chromatin immunoprecipitation assays (top inset) was generated. The effects of exogenously expressed caSTAT5a, WT STAT5a, or STAT5a Y-F MUT on LEF-1 autoregulation were then tested in HEK293 cells co-transfected with the LEF-1 reporter construct. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (B) LEF-1 mRNA expression in HEK293 cells transfected as described in panel A. mRNA was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (C) The effects of caSTAT5a, WT STAT5a, or STAT5a Y-F MUT on the LEF-1–mediated activation of a TOP promoter construct containing 4 LEF-1/TCFs binding sites were analyzed in HEK 293 cells co-transfected with the TOP construct and the corresponding expression plasmids by measuring the luciferase activity of the TOP promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (D,E) The effects of caSTAT5a on the LEF-1–mediated activation of C/EBPα and cyclin D1 promoters were analyzed in HEK 293 cells co-transfected with cyclin D1 (D) or C/EBPα (E) reporter gene constructs and the corresponding expression plasmids. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (F) Three STAT5 binding sites were deleted in the LEF-1 gene promoter (top inset). caSTAT5a-dependent inhibition of LEF-1 promoter autoregulation was determined in HEK293 cells co-transfected with the promoter construct containing mutated STAT5 binding sites and the corresponding expression plasmids by assessing the luciferase activity of the mutated LEF-1 promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (**P < .01). (G) Five LEF-1 binding sites were deleted in the LEF-1 promoter (top inset) caSTAT5a-dependent inhibition of LEF-1 promoter autoregulation was determined in HEK293 cells co-transfected with the promoter construct containing mutated LEF-1 binding sites and the corresponding expression plasmids by assessing the luciferase activity of the mutated LEF-1 promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate.
caSTAT5a inhibits the transcriptional activity of the LEF-1 promoter. (A) A LEF-1 reporter construct containing a 4000-bp upstream region of the LEF-1 gene harboring 3 binding sites identified in chromatin immunoprecipitation assays (top inset) was generated. The effects of exogenously expressed caSTAT5a, WT STAT5a, or STAT5a Y-F MUT on LEF-1 autoregulation were then tested in HEK293 cells co-transfected with the LEF-1 reporter construct. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (B) LEF-1 mRNA expression in HEK293 cells transfected as described in panel A. mRNA was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (C) The effects of caSTAT5a, WT STAT5a, or STAT5a Y-F MUT on the LEF-1–mediated activation of a TOP promoter construct containing 4 LEF-1/TCFs binding sites were analyzed in HEK 293 cells co-transfected with the TOP construct and the corresponding expression plasmids by measuring the luciferase activity of the TOP promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (D,E) The effects of caSTAT5a on the LEF-1–mediated activation of C/EBPα and cyclin D1 promoters were analyzed in HEK 293 cells co-transfected with cyclin D1 (D) or C/EBPα (E) reporter gene constructs and the corresponding expression plasmids. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (F) Three STAT5 binding sites were deleted in the LEF-1 gene promoter (top inset). caSTAT5a-dependent inhibition of LEF-1 promoter autoregulation was determined in HEK293 cells co-transfected with the promoter construct containing mutated STAT5 binding sites and the corresponding expression plasmids by assessing the luciferase activity of the mutated LEF-1 promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (**P < .01). (G) Five LEF-1 binding sites were deleted in the LEF-1 promoter (top inset) caSTAT5a-dependent inhibition of LEF-1 promoter autoregulation was determined in HEK293 cells co-transfected with the promoter construct containing mutated LEF-1 binding sites and the corresponding expression plasmids by assessing the luciferase activity of the mutated LEF-1 promoter. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate.
We also analyzed the effects of caSTAT5a on LEF-1–mediated transactivation using promoter reporter constructs of the LEF-1 target genes cyclin D1 and C/EBPα as well as the TOP/FOP flash reporter system (Figure 2C-E and supplemental Figure 1A-C). The TOP luciferase reporter construct contains 4 binding sites for LEF-1/TCFs, whereas these binding sites are absent in the FOP construct. Consistent with caSTAT5a inhibition of LEF-1 autoregulation, co-transfection of caSTAT5a and LEF-1 resulted in a substantial reduction in TOP promoter activity, but not FOP promoter activity. Again, WT STAT5a and Y-F STAT5a MUT had no inhibitory effects on LEF-1–mediated TOP promoter activation (Figure 2C). caSTAT5a also inhibited LEF-1–triggered activation of cyclin D1 and C/EBPα promoter constructs (Figure 2D-E). Both cyclin D1 and C/EBPα are direct target genes of LEF-1. A promoter construct of the β-casein gene (pZZ1), known to be a target gene of STAT5, was used as a positive control for STAT5 action (supplemental Figure 1D).
We next evaluated whether caSTAT5a inhibited LEF-1 activities through direct binding to the LEF-1 gene promoter using a LEF-1 gene promoter construct in which STAT5 binding sites were deleted. Intriguingly, caSTAT5a inhibited LEF-1–dependent autoregulation even if STAT5 binding sites were mutated (Figure 2F), suggesting that the STAT5a-dependent reduction in LEF-1 promoter activity did not require binding to the LEF-1 gene promoter. However, introduction of mutations in the LEF-1 binding sites almost completely abolished the activating effects of exogenously expressed LEF-1 on its own promoter and eliminated the inhibitory effect of co-transfected caSTAT5a (Figure 2G).
STAT5 protein interacts with LEF-1 protein
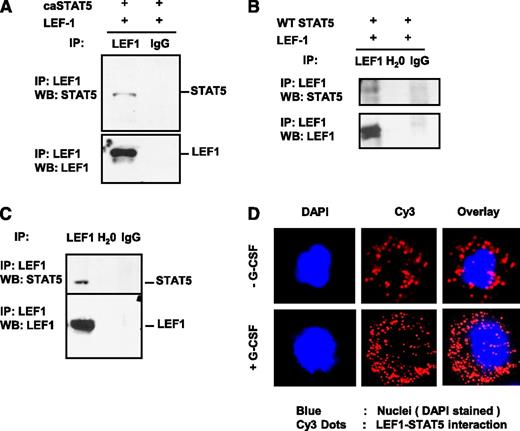
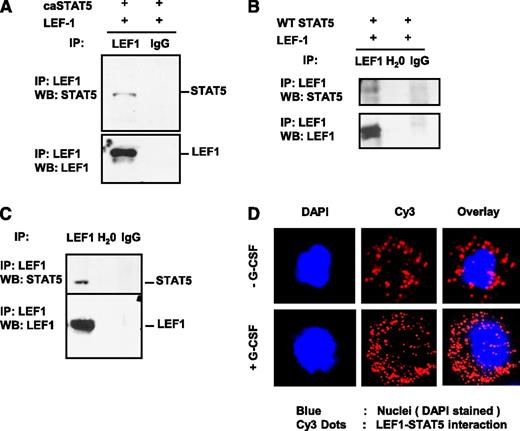
That caSTAT5a-dependent inhibition of LEF-1 autoregulation depended on the integrity of LEF-1 promoter binding sites, but not STAT5 binding sites, suggests that STAT5a may act through interaction with LEF-1 protein. To investigate this possibility, we immunoprecipitated LEF-1 from lysates of HEK293 cells co-transfected with LEF-1 and caSTAT5a or WT STAT5a using an anti–LEF-1 antibody, and probed immunoprecipitates by western blot analysis using anti–LEF-1 and anti-STAT5a antibodies. We found that caSTAT5a protein interacted directly with LEF-1 protein in co-transfected HEK293 cells (Figure 3A). Co-transfected WT STAT5a also interacted with LEF-1 protein, albeit to a lesser extent (Figure 3B).
STAT5 protein interacts with LEF-1 protein. Whole-cell lysates of HEK293 cells co-transfected with expression plasmids for LEF-1 and caSTAT5a complementary DNA (A) or WT STAT5a (B) were immunoprecipitated (IP) with rabbit monoclonal anti–LEF-1 antibody (IP-antibody) and were further analyzed by western blotting (WB) using a mouse monoclonal anti–LEF-1 or mouse monoclonal anti-STAT5a antibody. Representative images are shown. (C) Jurkat cell lysates were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to WB analysis with monoclonal anti–LEF-1 antibody or polyclonal anti-STAT5a antibody. Lysates of Jurkat cells used for immunoprecipitation were included as a positive control. Representative WB images are shown. (D) The interaction of endogenous LEF-1 and STAT5a proteins in CD34+ cells from healthy individuals incubated with or without 10 ng/mL of G-CSF was detected using a Duolink In Situ Proximity-Ligation Assay. (Left) DAPI-stained nuclei; (middle) Cy3 dots corresponding to LEF-1–STAT5a interaction; (right) DAPI/Cy3 overlay with red dots corresponding to LEF-1–STAT5a interaction. Ig, immunoglobulin.
STAT5 protein interacts with LEF-1 protein. Whole-cell lysates of HEK293 cells co-transfected with expression plasmids for LEF-1 and caSTAT5a complementary DNA (A) or WT STAT5a (B) were immunoprecipitated (IP) with rabbit monoclonal anti–LEF-1 antibody (IP-antibody) and were further analyzed by western blotting (WB) using a mouse monoclonal anti–LEF-1 or mouse monoclonal anti-STAT5a antibody. Representative images are shown. (C) Jurkat cell lysates were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to WB analysis with monoclonal anti–LEF-1 antibody or polyclonal anti-STAT5a antibody. Lysates of Jurkat cells used for immunoprecipitation were included as a positive control. Representative WB images are shown. (D) The interaction of endogenous LEF-1 and STAT5a proteins in CD34+ cells from healthy individuals incubated with or without 10 ng/mL of G-CSF was detected using a Duolink In Situ Proximity-Ligation Assay. (Left) DAPI-stained nuclei; (middle) Cy3 dots corresponding to LEF-1–STAT5a interaction; (right) DAPI/Cy3 overlay with red dots corresponding to LEF-1–STAT5a interaction. Ig, immunoglobulin.
To investigate the interaction between endogenous LEF-1 and STAT5a proteins, we immunoprecipitated LEF-1 from lysates of human Jurkat T lymphoblastic cells, which constitutively express high levels of both STAT5a and LEF-1 proteins. Jurkat cell lysates were immunoprecipitated with an anti–LEF-1 antibody and the immunoprecipitates were subjected to western blot analysis with an anti-STAT5a antibody. As shown in Figure 3C, endogenous STAT5a protein co-immunoprecipitated with endogenous LEF-1 protein.
Using a Duolink in situ Proximity Ligation Assay, we also detected the interaction of endogenous LEF-1 and STAT5a proteins in CD34+ cells of healthy individuals, an interaction that was substantially increased by treatment of cells with G-CSF (Figure 3D).
caSTAT5a–LEF-1 interactions lead to degradation of LEF-1 protein with subsequent inhibition of autoregulation of LEF-1 mRNA expression
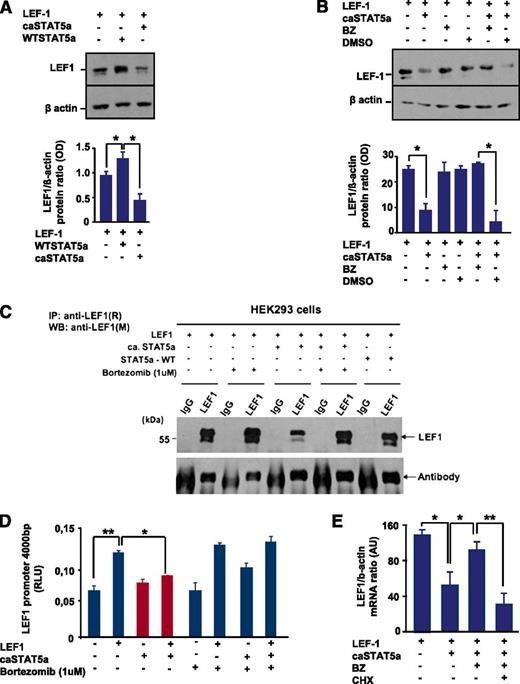
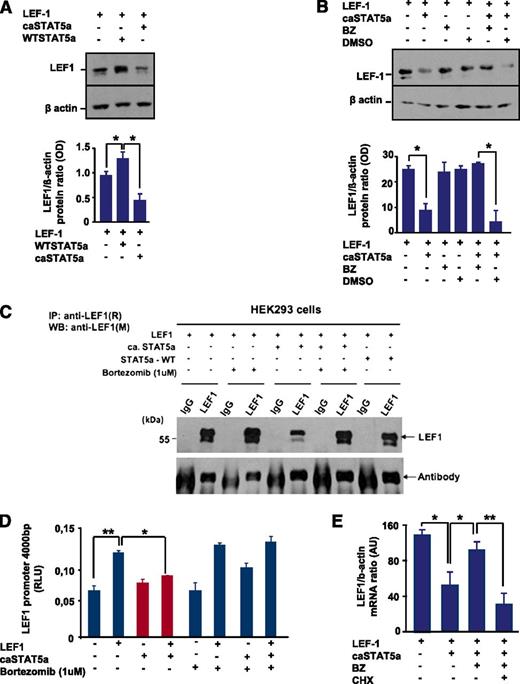
Interestingly, we found a decrease in LEF-1 protein levels after co-transfection of HEK293 cells with LEF-1 and caSTAT5a complementary DNAs compared with cells transfected with LEF-1 alone (Figure 4A). WT STAT5a binds to LEF-1 protein, but this binding had no effect on LEF-1 protein expression (Figure 4A,C). To evaluate the mechanism of the caSTAT5a-dependent reduction in LEF-1 protein levels, we treated transfected 293 cells with bortezomib, a potent and selective inhibitor of the ubiquitin-proteasome pathway. We found that treatment with bortezomib protected LEF-1 protein from caSTAT5a-dependent degradation (Figure 4B-C). Treatment with bortezomib alone did not substantially affect LEF-1, STAT5a, or phospho-STAT5a levels (Figure 4B-C and data not shown), but restored STAT5a-dependent inhibition of LEF-1 transcriptional activity in reporter gene assays using the LEF-1 promoter construct (Figure 4D).
caSTAT5a, but not WT STAT5a, downregulates the expression of LEF-1 protein, an effect that is rescued by bortezomib. (A) (Top) HEK293 cells were transfected with an LEF-1 expression plasmid with our without co-transfection of caSTAT5a or WT STAT5a. Whole-cell lysates were analyzed for LEF-1 and β-actin protein expression by western blotting. All samples were processed in a similar fashion, as described in Materials and Methods. Representative images are shown. (Bottom) Bar graphs indicate the ratio between the optical density of protein bands of LEF-1 protein to that of the housekeeping protein, β-actin. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05). (B) (Top) HEK293 cells were transfected with an LEF-1 expression plasmid, with our without co-transfection of caSTAT5a, and subsequently treated with 1 μg/μL of bortezomib (BZ) or dimethylsulfoxide. Whole-cell lysates were analyzed for LEF-1 and β-actin protein expression by western blotting. All samples were processed in a similar fashion, as described in Materials and Methods. Representative images are shown. (Bottom) Bar graphs indicate the ratio between the optical density of protein bands of LEF-1 protein to that of the housekeeping protein, β-actin. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05). (C) HEK293 cells were transfected with an LEF-1 expression plasmid, with our without co-transfection of caSTAT5a or WT STAT5a, and subsequently treated with 1 μg/μL BZ. Lysates of transfected cells were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to western blot analysis with a monoclonal anti–LEF-1 antibody. The IgG antibody band was used as a loading control. Representative western blot images are shown. (D) LEF-1 mRNA expression in HEK293 cells transfected as described in panel C. mRNA was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (E) HEK293 cells were transfected with an LEF-1 expression plasmid, with or without co-transfection of caSTAT5a or WT STAT5a, and subsequently incubated with 1 μg/mL of BZ alone or in combination with 1 μg/mL of cycloheximide (CHX). After 24 hours, mRNA was isolated and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05, **P < .01).
caSTAT5a, but not WT STAT5a, downregulates the expression of LEF-1 protein, an effect that is rescued by bortezomib. (A) (Top) HEK293 cells were transfected with an LEF-1 expression plasmid with our without co-transfection of caSTAT5a or WT STAT5a. Whole-cell lysates were analyzed for LEF-1 and β-actin protein expression by western blotting. All samples were processed in a similar fashion, as described in Materials and Methods. Representative images are shown. (Bottom) Bar graphs indicate the ratio between the optical density of protein bands of LEF-1 protein to that of the housekeeping protein, β-actin. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05). (B) (Top) HEK293 cells were transfected with an LEF-1 expression plasmid, with our without co-transfection of caSTAT5a, and subsequently treated with 1 μg/μL of bortezomib (BZ) or dimethylsulfoxide. Whole-cell lysates were analyzed for LEF-1 and β-actin protein expression by western blotting. All samples were processed in a similar fashion, as described in Materials and Methods. Representative images are shown. (Bottom) Bar graphs indicate the ratio between the optical density of protein bands of LEF-1 protein to that of the housekeeping protein, β-actin. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05). (C) HEK293 cells were transfected with an LEF-1 expression plasmid, with our without co-transfection of caSTAT5a or WT STAT5a, and subsequently treated with 1 μg/μL BZ. Lysates of transfected cells were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to western blot analysis with a monoclonal anti–LEF-1 antibody. The IgG antibody band was used as a loading control. Representative western blot images are shown. (D) LEF-1 mRNA expression in HEK293 cells transfected as described in panel C. mRNA was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are expressed as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05, **P < .01). (E) HEK293 cells were transfected with an LEF-1 expression plasmid, with or without co-transfection of caSTAT5a or WT STAT5a, and subsequently incubated with 1 μg/mL of BZ alone or in combination with 1 μg/mL of cycloheximide (CHX). After 24 hours, mRNA was isolated and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05, **P < .01).
To evaluate whether caSTAT5a blocks autoregulation of LEF-1, we co-transfected HEK293 cells with LEF-1 and caSTAT5a expression plasmids and found that caSTAT5a did indeed inhibit LEF-1 mRNA expression (Figure 4E). We additionally treated cells with bortezomib and demonstrated that treatment with bortezomib restored caSTAT5a-dependent inhibition of LEF-1 mRNA synthesis (Figure 4E). Furthermore, we found that treatment with the protein synthesis inhibitor cycloheximide blocked the ability of bortezomib to induce LEF-1 mRNA (Figure 4E). Collectively, these data suggest that caSTAT5a inhibits the autoregulatory effects of LEF-1 protein on LEF-1 mRNA and protein synthesis.
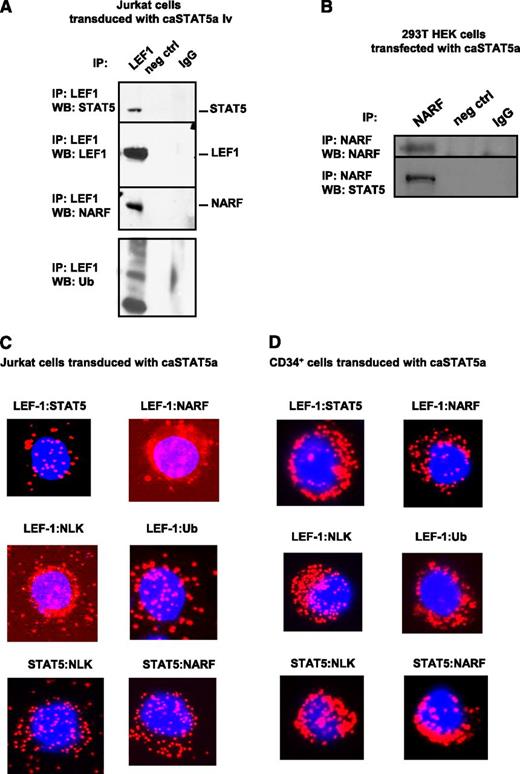
Hyperactivated STAT5a recruits the E3 ubiquitin ligase NARF to LEF-1 protein, inducing LEF-1 ubiquitination
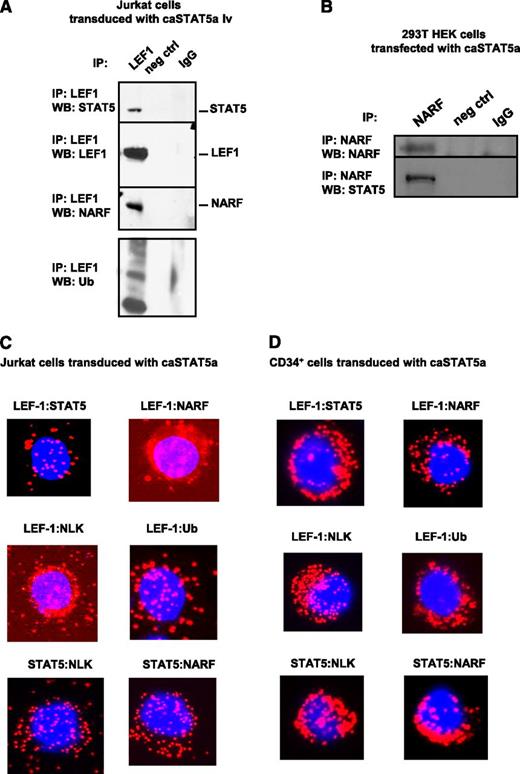
We further investigated the mechanism of caSTAT5a-dependent degradation of LEF-1 protein. G-CSF has been previously shown to activate NLK,33 which recruits the E3 ubiquitin ligase NARF to LEF-1 protein, leading to LEF-1 ubiquitination and degradation.32 Moreover, activated STAT3 has been shown to interact with and activate NLK.34 Therefore, we evaluated whether caSTAT5a also interacts with NLK, leading to the recruitment of NARF to LEF-1 and subsequent LEF-1 ubiquitination. Indeed, we found that NARF and ubiquitin immunoprecipitated with LEF-1 in Jurkat cells transduced with caSTAT5a (Figure 5A). Interaction between caSTAT5 and NARF was confirmed by coimmunoprecipitation using lysates of 293T cells transfected with caSTAT5 (Figure 5B). A Duolink analysis further revealed interactions between NARF and LEF-1, NLK and LEF-1, ubiquitin and LEF-1, NLK and STAT5, and NARF and STAT5 in Jurkat cells and CD34+ cells after transduction with caSTAT5a (Figure 5C-D).
caSTAT5a-NLK-NARF–mediated ubiquitination of LEF-1. Interactions between LEF-1, STAT5, NARF, and NLK proteins as well as LEF-1 ubiquitination: (A) lysates of Jurkat cells transduced with caSTAT5A were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to western blot analysis with anti–LEF-1, anti-STAT5, anti-NARF, anti-NLK, or anti-ubiquitin antibodies. Representative western blot images are shown; (B) lysates of 293T HEK cells transfected with caSTAT5A were immunoprecipitated with anti-NARF antibody and the immunoprecipitates were subjected to western blot analysis with anti-NARF or anti-STAT5 antibodies; representative western blot images are shown. (C-D) Interactions between LEF-1 and STAT5, LEF-1 and NARF, LEF-1 and NLK, LEF-1 and ubiquitin, STAT5 and NLK, and STAT5 and NARF were detected in (C) Jurkat cells and (D) CD34+ cells treated with G-CSF using Duolink. DAPI, stained nuclei; Cy3 red dots, protein interactions.
caSTAT5a-NLK-NARF–mediated ubiquitination of LEF-1. Interactions between LEF-1, STAT5, NARF, and NLK proteins as well as LEF-1 ubiquitination: (A) lysates of Jurkat cells transduced with caSTAT5A were immunoprecipitated with a polyclonal anti–LEF-1 antibody and the immunoprecipitates were subjected to western blot analysis with anti–LEF-1, anti-STAT5, anti-NARF, anti-NLK, or anti-ubiquitin antibodies. Representative western blot images are shown; (B) lysates of 293T HEK cells transfected with caSTAT5A were immunoprecipitated with anti-NARF antibody and the immunoprecipitates were subjected to western blot analysis with anti-NARF or anti-STAT5 antibodies; representative western blot images are shown. (C-D) Interactions between LEF-1 and STAT5, LEF-1 and NARF, LEF-1 and NLK, LEF-1 and ubiquitin, STAT5 and NLK, and STAT5 and NARF were detected in (C) Jurkat cells and (D) CD34+ cells treated with G-CSF using Duolink. DAPI, stained nuclei; Cy3 red dots, protein interactions.
Restoration of LEF-1 expression by treatment of CN CD34+ cells with bortezomib or transduction with LEF-1 lentivirus restores in vitro G-CSF–triggered granulocytic differentiation
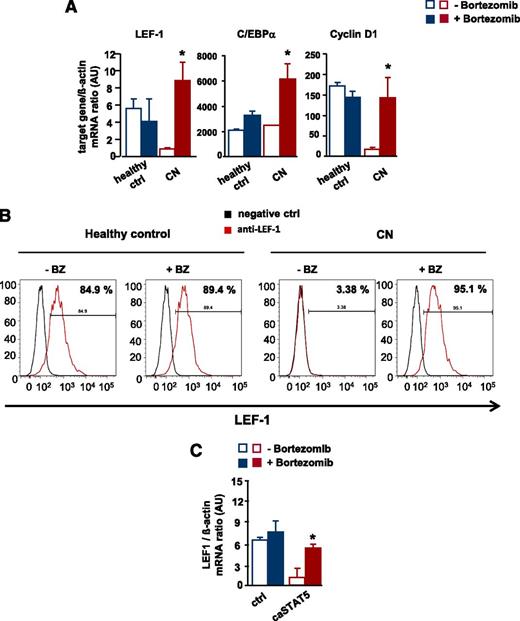
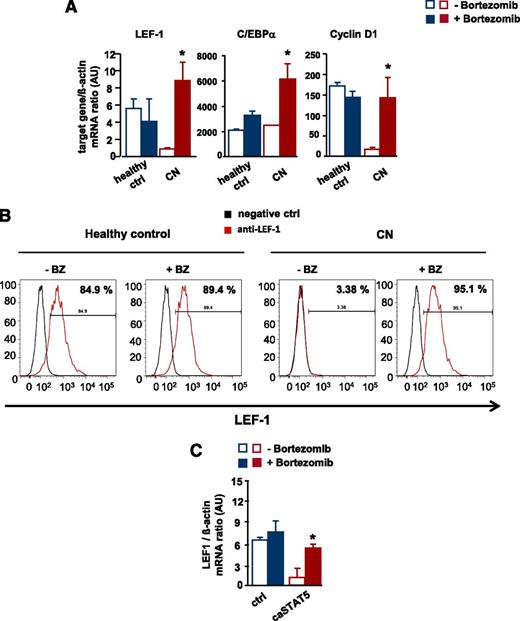
To address the significance of the bortezomib-mediated reversal of the STAT5-dependent reduction in LEF-1 mRNA and protein levels, we analyzed in vitro G-CSF–triggered granulocytic differentiation in CD34+ cells from 2 CN patients and healthy individuals in the presence and absence of bortezomib. Bortezomib markedly upregulated LEF-1 mRNA and protein levels compared with untreated control groups and increased mRNA expression of the LEF-1 target genes C/EBPα and cyclin D1 (Figure 6A-B and supplemental Figure 2A-B).
Restoration of defective LEF-1 expression and activity by treatment of CN CD34+ progenitors with bortezomib. In vitro G-CSF–triggered granulocytic differentiation of CD34+ cells from CN patients and healthy individuals was performed in the presence or absence of bortezomib (10 nM), as described in Materials and Methods. (A) mRNA expression of LEF-1 and the LEF-1 target genes C/EBPα and cyclin D1 in studied groups, as assessed by RT-PCR. LEF-1, C/EBPα, and cyclin D1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (B) Intracellular expression levels of LEF-1 protein detected using FACS analysis, as described in “Materials and methods”. Representative histograms as well as percentages of positive cells are depicted. (C) CD34+ cells from healthy individuals were transduced with caSTAT5a or ctrl rv constructs and incubated with or without bortezomib (10 nM). After 26 hours of treatment, cells were sorted and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05).
Restoration of defective LEF-1 expression and activity by treatment of CN CD34+ progenitors with bortezomib. In vitro G-CSF–triggered granulocytic differentiation of CD34+ cells from CN patients and healthy individuals was performed in the presence or absence of bortezomib (10 nM), as described in Materials and Methods. (A) mRNA expression of LEF-1 and the LEF-1 target genes C/EBPα and cyclin D1 in studied groups, as assessed by RT-PCR. LEF-1, C/EBPα, and cyclin D1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (*P < .05, **P < .01). (B) Intracellular expression levels of LEF-1 protein detected using FACS analysis, as described in “Materials and methods”. Representative histograms as well as percentages of positive cells are depicted. (C) CD34+ cells from healthy individuals were transduced with caSTAT5a or ctrl rv constructs and incubated with or without bortezomib (10 nM). After 26 hours of treatment, cells were sorted and LEF-1 mRNA expression was measured by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05).
In addition, treatment with bortezomib restored caSTAT5a-dependent inhibition of LEF-1 mRNA expression in CD34+ cells from healthy individuals transduced with caSTAT5a (Figure 6C).
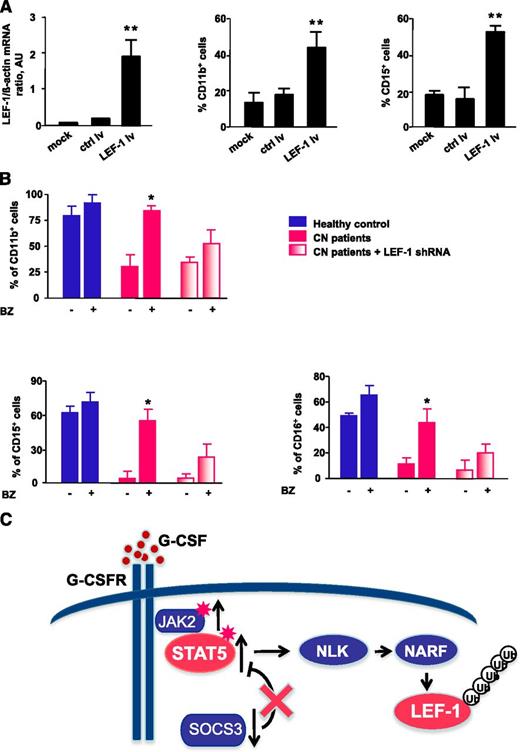
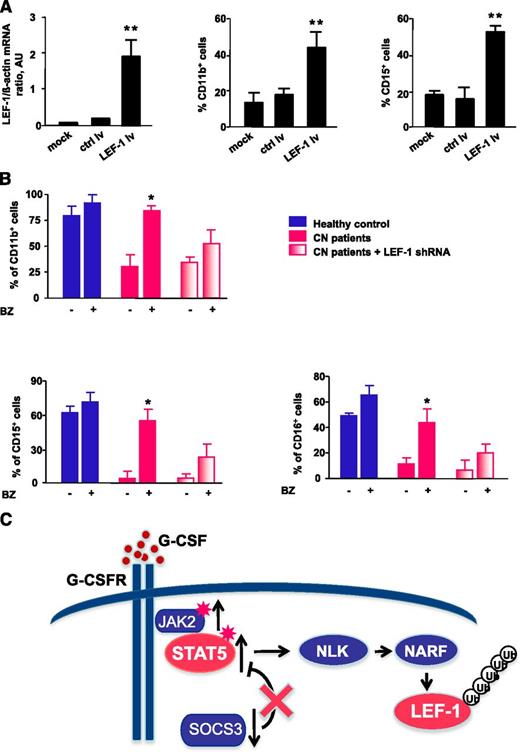
Transduction of CD34+ cells from CN patients with a lentivirus containing an LEF-1 expression construct led to restoration of LEF-1 mRNA expression in association with normalization of the defective G-CSF–triggered granulocytic differentiation, as assessed by surface expression of the granulocytic-specific markers CD11b, CD15, and CD16 (Figure 7A and data not shown).
Treatment of CN CD34+ progenitors with bortezomib or transduction of cells with LEF-1 lv promotes in vitro granulocytic differentiation. (A) CD34+ cells from CN patients (n = 2) were transduced with LEF-1 lv or ctrl lv, and granulocytic differentiation was induced as described in Materials and Methods; untransduced cells (controls) were cultured under the same conditions. Expression of LEF-1 mRNA and granulocytic (CD15) and myeloid (CD11b) surface markers as assessed on day 7 of culture. Cell surface marker expression was measured by FACS; representative histograms are presented. mRNA expression were assessed by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (**P < .01). (B) In vitro G-CSF–triggered granulocytic differentiation of CD34+ cells from 2 CN patients and 2 healthy individuals was performed in the presence or absence of bortezomib (10 nM), as described in Materials and Methods. Some cells from CN patients were transduced with lentivirus-based LEF-1 shRNA constructs. Surface expression of the granulocyte-specific markers, CD11b, CD15, and CD16, were assessed using FACS. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). (C) Schematic representation of the caSTAT5a-dependent degradation of LEF-1 protein in myeloid cells of CN patients. In CN patients, daily treatment with high pharmacological doses with G-CSF results in hyperactivation of STAT5a by phosphorylation owing to elevated JAK2 and diminished SOCS3 expression levels. Hyperactivated STAT5 activates NLK, promoting NLK–NARF interactions, and leading to recruitment of the NARF-NLK complex to LEF-1 protein and subsequent LEF-1 ubiquitination and degradation.
Treatment of CN CD34+ progenitors with bortezomib or transduction of cells with LEF-1 lv promotes in vitro granulocytic differentiation. (A) CD34+ cells from CN patients (n = 2) were transduced with LEF-1 lv or ctrl lv, and granulocytic differentiation was induced as described in Materials and Methods; untransduced cells (controls) were cultured under the same conditions. Expression of LEF-1 mRNA and granulocytic (CD15) and myeloid (CD11b) surface markers as assessed on day 7 of culture. Cell surface marker expression was measured by FACS; representative histograms are presented. mRNA expression were assessed by qRT-PCR. LEF-1 mRNA levels were normalized to those of β-actin and are presented as AUs. Data represent means ± SD and are derived from 3 independent experiments, each in triplicate (**P < .01). (B) In vitro G-CSF–triggered granulocytic differentiation of CD34+ cells from 2 CN patients and 2 healthy individuals was performed in the presence or absence of bortezomib (10 nM), as described in Materials and Methods. Some cells from CN patients were transduced with lentivirus-based LEF-1 shRNA constructs. Surface expression of the granulocyte-specific markers, CD11b, CD15, and CD16, were assessed using FACS. Data represent means ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). (C) Schematic representation of the caSTAT5a-dependent degradation of LEF-1 protein in myeloid cells of CN patients. In CN patients, daily treatment with high pharmacological doses with G-CSF results in hyperactivation of STAT5a by phosphorylation owing to elevated JAK2 and diminished SOCS3 expression levels. Hyperactivated STAT5 activates NLK, promoting NLK–NARF interactions, and leading to recruitment of the NARF-NLK complex to LEF-1 protein and subsequent LEF-1 ubiquitination and degradation.
We further analyzed whether bortezomib treatment enabled G-CSF–triggered terminal granulocytic differentiation of CD34+ cells from CN patients. Indeed, treatment with bortezomib overcame the maturation block evident in CN progenitors, whereas CN progenitors not treated with bortezomib remained defective for G-CSF–dependent granulopoiesis (Figure 7B). To evaluate whether bortezomib induced granulopoiesis in CN progenitors via restoration of LEF-1 expression, we transduced CD34+ cells from CN patients with small (inhibitory) hairpin RNA (shRNA) against LEF-1 and evaluated myeloid differentiation in vitro. Differentiation was reduced in bortezomib-treated CD34+ cells transduced with LEF-1 shRNA (Figure 7B), suggesting that bortezomib acts through upregulation of LEF-1 and increased expression of LEF-1 target genes. The residual granulocytic differentiation observed in CD34+ cells transduced with LEF-1 shRNA could be explained by the presence of residual LEF-1 levels (ie, incomplete LEF-1 downregulation) or by the activation of another LEF-1–independent pathway of granulopoiesis by bortezomib.
Discussion
In the present study, we found a significant and sustained elevation in the levels of phospho-STAT5 in CD34+ cells from CN patients compared with those from healthy individuals. Phospho-STAT5 levels were even higher in a CN patient who subsequently developed AML. Because “steady-state” granulopoiesis via C/EBPα is defective in CN patients,27 daily treatment with high doses of G-CSF are required to support “emergency” granulopoiesis via C/EBPβ, which is not affected in CN.5 This is the case regardless of the mutational status (eg, ELA2 or HAX1 mutations) of the CN patient. High levels of G-CSF act as a persistent trigger for activation of JAK kinases associated with G-CSFR signaling.40 Hyperactivated JAK2 in combination with diminished expression of SOCS3 could lead to elevated levels of phospho-STAT5 in myeloid cells of CN patients. Additionally, ELA2 mutations in myeloid cells of CN patients lead to cytoplasmic accumulation of elastase protein, followed by activation of the unfolded protein response and endoplasmic reticulum stress.41 Endoplasmic reticulum stress/unfolded protein response is also associated with induction of the JAK2/STAT5 signaling pathway42 ; therefore, the unfolded protein response could also induce sustained phosphorylation of STAT5 in CN patients.
Hyperactivated STAT5 has been implicated in various hematological malignancies, including AML, and in myeloproliferative disorders.16 Our data might indicate that leukemic transformation secondary to CN is, in part, a consequence of enhanced activation of STAT5.
Previously, we reported that LEF-1 expression was defective or even absent in myeloid cells of CN patients.27 In line with this, we also found that LEF-1 levels were undetectable in CD34+ cells from the CN/AML patient, further confirming that these cells lack differentiation potential and are more susceptible to the G-CSF–induced proliferative response, effects that are attributable to prolonged expression of hyperphosphorylated STAT5a. Hyperactivated STAT5a is also known to impair myelopoiesis, consistent with suppression of C/EBPα.14,15 Our investigation of the possible association between sustained levels of hyperactivated STAT5a and downregulation of LEF-1 and its target gene, C/EBPα, showed that overexpression of caSTAT5a, but not WT STAT5a or STAT5a Y-F MUT, in CD34+ hematopoietic progenitors has an inhibitory effect on LEF-1 and C/EBPα. However, we cannot exclude the possibility that hyperactivated STAT5a could also function in the partial granulopoietic differentiation that takes place in CN patients via activation of C/EBPβ-dependent emergency granulopoiesis. C/EBPβ is known to interact with STAT5 on the promoter,43 and C/EBPβ levels are elevated in CN.5 We did not find any effects of caSTAT5a on erythropoiesis, as has been suggested previously.13 This apparent discrepancy could be explained by differences in the protocols used for transduction and culture of CD34+ cells.
STAT5-mediated gene regulation mainly depends on STAT5 transactivation potential, but several recent reports have emphasized the DNA binding-dependent and -independent repressive functions of STAT5.17,44 Reporter gene assays showed that LEF-1 promoter constructs containing mutated STAT5 binding sites retained caSTAT5a-dependent inhibition of LEF-1 autoregulation, suggesting that STAT5a-dependent repression of LEF-1 autoregulation is not mediated by direct DNA binding.
In the present study, we provide the first evidence for the interaction between STAT5a and LEF-1 proteins. LEF-1 is a known target of ubiquitination and its turnover in cells is regulated by the proteasome pathway.33,45 Intriguingly, we found that overexpression of caSTAT5a, but not WT STAT5a or STAT5a Y-F MUT, led to enhanced degradation of LEF-1 protein and, subsequently, to suppression of LEF-1–dependent autoregulation of the LEF-1 promoter and downregulation of the mRNA levels of LEF-1 and LEF-1 target genes. Under conditions of dysregulated signaling with concurrent hyperactivation of STAT5a, as occurs in CN, interactions between LEF-1 and STAT5 might be responsible for the recruitment of ubiquitin-protein ligases, which, in turn, would lead to elevated proteasomal degradation of LEF-1 protein. Indeed, we found that, in addition to LEF-1, other factors such as TCFs are known to bind and regulate the LEF-1 promoter. However, we found no STAT5a-dependent repression of the LEF-1 promoter if LEF-1 binding sites were mutated. This suggests that caSTAT5a inhibition of LEF-1 is closely related to disturbed LEF-1 autoregulation. Another possibility for the inhibitory action of activated STAT5a on the LEF-1 protein expression is via its induction of another gene that might then affect LEF1 stability.
Consistent with this, we found that treatment with the proteasome inhibitor bortezomib exerted protective effects against STAT5a-dependent LEF-1 protein degradation. Moreover, treatment with bortezomib restored caSTAT5a-dependent inhibition of LEF-1 mRNA and protein expression in HEK293 cells and led to significantly elevated levels of LEF-1 and LEF-1 target genes in CN patients. Bortezomib restored in vitro G-CSF–triggered granulocytic differentiation in association with elevated LEF-1 expression in CN patients. A recent study of bortezomib in the treatment of chronic myeloid leukemia showed that bortezomib alone has no effect on the levels of phosphorylated STAT5a, and little or no effect on STAT5/DNA binding.46 Therefore, bortezomib-induced restoration of granulocytic differentiation in CN is most likely to the result of inhibition of LEF-1 protein degradation. LEF-1 shRNA prevented bortezomib-induced granulocytic differentiation. Residual granulocytic differentiation in CD34+ cells transduced with LEF-1 shRNA could be explained by the presence of residual LEF-1 levels or by the activation of another LEF-1–independent pathway of granulopoiesis by bortezomib. In addition, treatment with cycloheximide, which inhibits protein translation, blocked the ability of bortezomib to induce LEF-1 mRNA, suggesting an effect of bortezomib on LEF-1 autoregulation. Effects similar to those of bortezomib treatment were observed after inhibition of STAT5 in CD34+ cells from CN patients with the pharmacological inhibitor pimozide, which restored LEF-1 expression in these cells. Therefore, we conclude that hyperactivated STAT5a, which is a downstream effector of the G-CSF signaling pathway, not only binds to LEF-1 protein, but also activates NLK. Activated NLK recruits NARF to LEF-1 protein and thereby supports formation of multiprotein complexes consisting of STAT5a, NLK, NARF, and LEF-1. This results in the ubiquitination and degradation of LEF-1 protein, leading to the diminished binding of LEF-1 protein to the LEF-1 promoter and abolished LEF-1 autoregulation (Figure 7C). Bortezomib prevents degradation of the LEF-1 protein and thereby restores LEF-1 autoregulation. This study opens novel avenues for potential clinical applications of the proteasome inhibitor bortezomib as a cellular differentiation agent for use in combination with G-CSF to treat some CN patients. Because side effects of long-term bortezomib treatment have been reported (eg, in myeloma patients), carefully designed, precise protocols of bortezomib therapy, for instance using reduced doses in combination with low doses of G-CSF, should be considered for applications in CN patients.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank K. Cherkaoui, A. Müller Brechlin, and M. Reuter for the excellent technical assistance; C. Zeidler and the physicians within the Severe Chronic Neutropenia International Registry for providing patients material; and to the study subjects for their cooperation.
Authorship
Contribution: K.W. and J.S. made initial observations, designed the experiments, analyzed the data, supervised experimentation, and wrote the manuscript; K.G. and I.K. performed the main experiments; O.K. performed part of the experiments with CD34+ cells; J.M. made the STAT5 constructs; M.A.S.M. suggested studying caSTAT5 in CN patients; and C.Z. provided patient materials.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Julia Skokowa, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: skokowa.julia@mh-hannover.de; and Karl Welte, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: Welte.Karl.H@mh-hannover.de.
References
Author notes
K.G. and I.K. contributed equally to this study.