Abstract
Most heritable anemias are caused by mutations in genes encoding globins, red blood cell (RBC) membrane proteins, or enzymes in the glycolytic and hexose monophosphate shunt pathways. A less common class of genetic anemia is caused by mutations that alter the functions of erythroid transcription factors (TFs). Many TF mutations associated with heritable anemia cause truncations or amino acid substitutions, resulting in the production of functionally altered proteins. Characterization of these mutant proteins has provided insights into mechanisms of gene expression, hematopoietic development, and human disease. Mutations within promoter or enhancer regions that disrupt TF binding to essential erythroid genes also cause anemia and heritable variations in RBC traits, such as fetal hemoglobin content. Defining the latter may have important clinical implications for de-repressing fetal hemoglobin synthesis to treat sickle cell anemia and β thalassemia. Functionally important alterations in genes encoding TFs or their cognate cis elements are likely to occur more frequently than currently appreciated, a hypothesis that will soon be tested through ongoing genome-wide association studies and the rapidly expanding use of global genome sequencing for human diagnostics. Findings obtained through such studies of RBCs and associated diseases are likely generalizable to many human diseases and quantitative traits.
Introduction
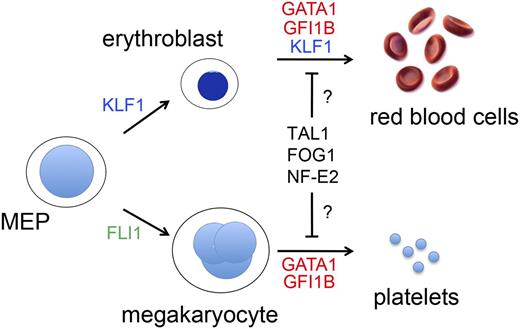
The most common hereditary forms of anemia arise from mutations in genes that encode globins, red blood cell (RBC) membrane proteins, or enzymes. However, mutations in RBC transcription factor (TF) genes, or the cis elements through which they bind to regulate gene expression, account for a rare, but informative subset of anemias. Megakaryocytes (MEGs) and RBCs derive from a common bipotential MEG-erythroid progenitor (MEP), and their development is regulated by several common TFs, such as GATA binding protein 1 (GATA1) (Figure 1). Accordingly, GATA1 gene mutations can present with anemia, thrombocytopenia, or both. Similarly, T-cell acute lymphoblastic leukemia 1 (TAL1), FOG family member 1 (FOG1), and nuclear factor, erythroid 2 (NF-E2) are erythro-megakaryocytic TFs that could hypothetically be altered in hereditary anemia/thrombocytopenia. In contrast, Fli-1 proto oncogene (FLI1) and Kruppel-like factor 1 (KLF1) drive mono-lineage production of MEGs and RBCs, respectively, and identified mutations in these genes affect only the single corresponding lineage.1,2 Extensive research over more than a decade has defined the biological properties of these and several other erythro-megakaryocytic TFs. Mutations that alter their structure and/or expression to cause human blood diseases provide valuable insights into the biology of hematopoiesis and hemoglobin switching. Further investigations into the mechanisms of these mutations should identify new strategies for treating more common congenital anemias, for example, by manipulating fetal hemoglobin expression in the setting of sickle cell anemia or β thalassemia.
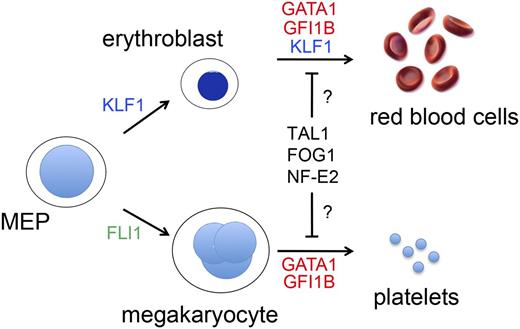
TFs that drive erythromegakaryopoiesis. RBCs and MEGs derive from the bipotential MEP. KLF1 and FLI1 regulate lineage determination of the MEP and subsequent erythroid or MEG maturation, respectively. GATA1, TAL1, FOG1, NF-E2, and GFI1B regulate the maturation of both lineages. Germ-line mutations in KLF1 (shown in blue) cause isolated congenital anemia, whereas mutations in the TFs indicated in red affect RBCs and MEGs. Candidate TFs for which no germ-line human mutations have yet been discovered to be associated with anemia or thrombocytopenia are shown in black. Deficiency of FLI1 (shown in green) is associated with thrombocytopenia, but not anemia in Paris-Trousseau syndrome.112
TFs that drive erythromegakaryopoiesis. RBCs and MEGs derive from the bipotential MEP. KLF1 and FLI1 regulate lineage determination of the MEP and subsequent erythroid or MEG maturation, respectively. GATA1, TAL1, FOG1, NF-E2, and GFI1B regulate the maturation of both lineages. Germ-line mutations in KLF1 (shown in blue) cause isolated congenital anemia, whereas mutations in the TFs indicated in red affect RBCs and MEGs. Candidate TFs for which no germ-line human mutations have yet been discovered to be associated with anemia or thrombocytopenia are shown in black. Deficiency of FLI1 (shown in green) is associated with thrombocytopenia, but not anemia in Paris-Trousseau syndrome.112
Etiologies of hereditary anemias
Hereditary anemia encompasses a wide spectrum of RBC disorders that typically present in infants and children. These disorders can be subclassified according to the RBC developmental stage most profoundly affected.
Shortened lifespan of circulating erythrocytes (hemolysis). This group includes the most common causes of hereditary anemia and is typically caused by defects in hemoglobin (sickle cell anemia and other hemoglobin variants), RBC membrane proteins (hereditary spherocytosis, elliptocytosis), or enzymatic pathways (glucose 6 phosphate dehyrogenase (G6PD] deficiency, glycolytic defects). Congenital dyserythropoietic anemia type II (CDA II) is also associated with early destruction of erythrocytes in the spleen.
Faulty maturation of erythroid precursors. Thalassemia syndromes are associated with impaired erythroid precursor maturation, termed “ineffective erythropoiesis.” The CDAs are associated with accumulation of erythroid precursors that form RBCs inefficiently.3 Affected erythroblasts often exhibit bizarre dysplastic morphologies including multinuclearity, internuclear bridging, and megaloblastic features. Historically, CDAs have been classified into 3 types (I, II, and III) based on the appearance of erythroid precursors and serological studies. Causal mutations for these classical CDAs have been identified in CDAN1 (CDA I), C15ORF41 (CDA I), SEC23B (CDA II), and KIF23 (CDA III).3,4 These genes do not encode typical hematopoietic TFs, although CDAN1 is reported to regulate the transport of histones5 and is implicated in the impaired localization of HP1α, a key component of heterochromatin.6 However, it is now clear that the CDAs are heterogeneous and some variant forms are caused by mutations in GATA1 and KLF1 (see below).
Reduced erythroid precursors (pure red cell aplasia/hypoplasia). The classical example is Diamond Blackfan anemia (DBA), a congenital anemia that typically presents in early childhood, often with associated developmental anomalies.7 DBA is genetically heterogeneous, with about two-thirds of cases caused by dominantly inherited heterozygous loss-of-function mutations in 1 of ≥12 large or small ribosomal subunit proteins. It is not certain how ribosomal protein haploinsufficiency causes selective defects in the accumulation of erythroid precursors. This may occur in part via perturbation of TF networks. For example, imbalanced ribosome assembly may activate p53, a TF that induces apoptosis and/or cell cycle arrest. In addition, GATA1 gene mutations are associated with an X-linked form of DBA8 (discussed in “How do defects in transcriptional regulation cause disease?”).
The majority of common hereditary anemias, including most hemoglobinopathies, membrane disorders, enzymopathies, and classical forms of CDA, are not caused by TF mutations. Once the clinical hematologist has ruled out these common causes, then mutations in erythroid or erythro-megakaryocytic TFs or their target cis elements become more likely and should be considered.
How do defects in transcriptional regulation cause disease?
Most essential erythroid or erythro-megakaryocytic TFs (so-called “master regulators”) activate or repress gene transcription globally to promote terminal maturation and suppress alternate lineages, respectively. Mutations that completely destroy the function of a TF by eliminating its expression or destabilizing the protein cause profound effects. In their homozygous state, these mutations are most likely embryonic lethal in humans, as predicted by numerous murine models. However, many mutations in human TF genes generate an altered, partially functional protein and are compatible with postnatal life, causing selective defects in gene expression and various clinical manifestations with a range of severity.
cis element mutations
TFs regulate gene expression by binding to specific cis element motifs in promoters and enhancers. Numerous human hematopoietic phenotypes are caused by polymorphisms or mutations in cis elements that recruit key TFs to DNA. For example, a subset of β thalassemias are caused by mutations that affect TF binding sites in the proximal β globin gene promoter and deletions of an upstream enhancer termed the locus control region.9 More recently, mutations in the DNA regulatory regions of numerous other erythroid genes have been shown to cause anemia.
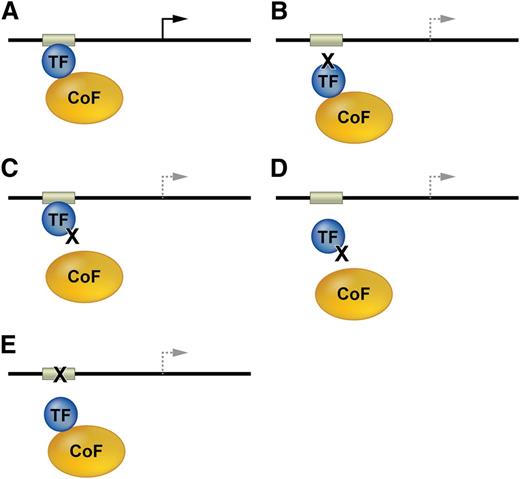
Mechanisms associated with mutations in TF genes or their cis elements in DNA regulatory regions are described generally in Figure 2, and specific salient examples are discussed in subsequent sections.
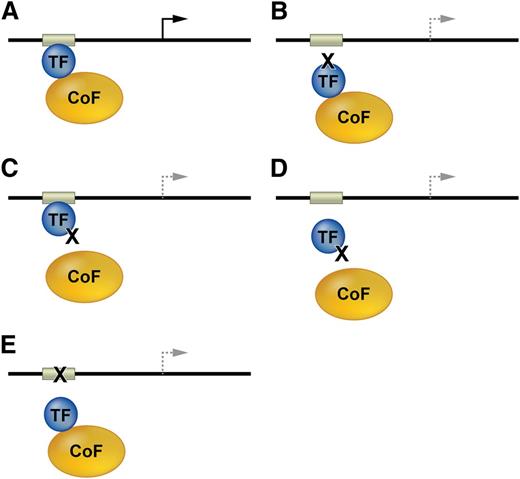
Mutations affecting transcription factor function. (A) TFs and bound cofactors (Cofs) regulate gene expression by binding cognate cis elements within DNA regulatory regions including promoters or enhancers (gray box). The arrow indicates gene transcription. (B) Mutations in the TF DNA binding domain inhibit TF interactions with some or all cis elements. The dashed lines indicate altered transcription. (C) TF mutations that inhibit Cof binding alter gene expression at some or all target genes. (D) Some Cof interactions are required for in vivo DNA occupancy by TFs. Thus, TF mutations outside of the DNA binding domain can inhibit gene occupancy. (E) Mutations in DNA cis elements can impair TF occupancy at a single site within one gene, thereby altering its transcription.
Mutations affecting transcription factor function. (A) TFs and bound cofactors (Cofs) regulate gene expression by binding cognate cis elements within DNA regulatory regions including promoters or enhancers (gray box). The arrow indicates gene transcription. (B) Mutations in the TF DNA binding domain inhibit TF interactions with some or all cis elements. The dashed lines indicate altered transcription. (C) TF mutations that inhibit Cof binding alter gene expression at some or all target genes. (D) Some Cof interactions are required for in vivo DNA occupancy by TFs. Thus, TF mutations outside of the DNA binding domain can inhibit gene occupancy. (E) Mutations in DNA cis elements can impair TF occupancy at a single site within one gene, thereby altering its transcription.
Mutations in erythro-megakaryocytic TFs required for RBC and platelet production.
Congenital anemia that is not believed to be caused by common mutations in genes encoding globins, membrane proteins, enzymes, or classical CDA should alert the clinical hematologist to the potential for mutations in genes encoding TFs. Accompanying thrombocytopenia may suggest a mutation in a TF that is shared by erythroid and MEG lineages. TF genes discovered to be mutated in human diseases are described in this section.
GATA1.
GATA1 is an X-linked TF that regulates maturation of multiple lineages including RBCs and MEGs.10 The full-length isoform contains 2 zinc fingers and an acidic N-terminal activation domain (NAD) (Figure 3A). The NAD is named by virtue of its ability to activate gene expression in transient reporter assays using heterologous cells.11 The C-terminal zinc finger (Cf) is responsible for the bulk of DNA binding activity, whereas the N-terminal zinc finger (Nf) stabilizes DNA binding at more complex GATA elements, including palindromic sites.12 The GATA1 Nf and Cf are similar in structure and both mediate interactions with multiple cofactor proteins. The GATA1 Nf specifically recruits the essential cofactor FOG1.13
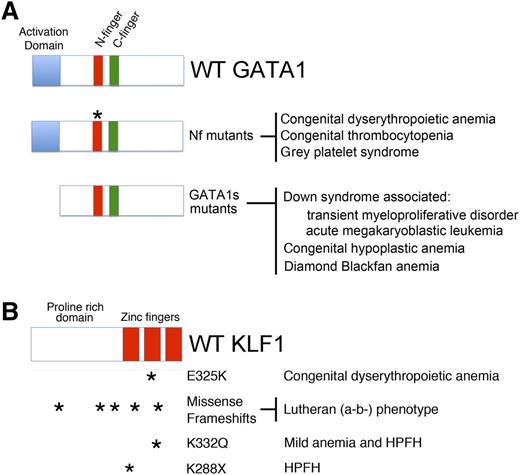
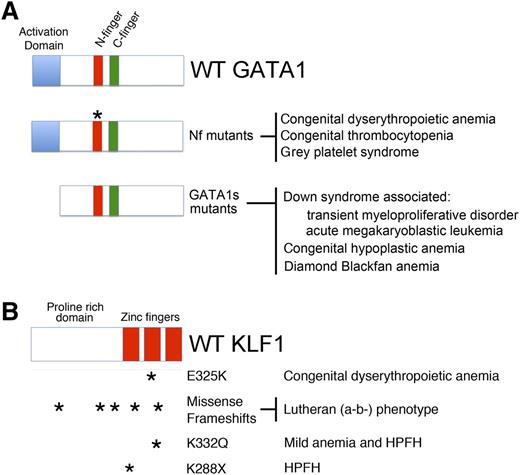
GATA1 and KLF1 domain modules, mutations, and associated diseases. (A) (Top) Three domains of GATA1 are shown: the N and C-terminal zinc fingers (Nf and Cf) and the NAD. (Middle) Mutations in the Nf (noted by asterisks) disrupt DNA binding or associations with the essential cofactors Friend of GATA1 (FOG-1) or TAL1 complex. These germ-line mutations cause a variety of inherited anemias and/or thrombocytopenias. (Bottom) Distinct mutations within exon 2 lead to predominant expression of GATA1s, which retains both zinc fingers, but is missing the NAD. In infants and young children with DS (trisomy 21), somatic mutations that favor GATA1s production cause TMD and AMKL. In euploid patients, germ-line GATA1s mutations are associated with congenital anemia, including DBA. (B) KLF1 includes an N-terminal proline-rich domain and 3 C-terminal zinc finger domains. Various missense and frameshift mutations (noted by asterisks) throughout the gene are associated with hereditary anemias, altered Lutheran blood group expression, and/or persistence of fetal hemoglobin expression. Note that E325K has a dominant negative effect. Adapted from Singleton et al.58
GATA1 and KLF1 domain modules, mutations, and associated diseases. (A) (Top) Three domains of GATA1 are shown: the N and C-terminal zinc fingers (Nf and Cf) and the NAD. (Middle) Mutations in the Nf (noted by asterisks) disrupt DNA binding or associations with the essential cofactors Friend of GATA1 (FOG-1) or TAL1 complex. These germ-line mutations cause a variety of inherited anemias and/or thrombocytopenias. (Bottom) Distinct mutations within exon 2 lead to predominant expression of GATA1s, which retains both zinc fingers, but is missing the NAD. In infants and young children with DS (trisomy 21), somatic mutations that favor GATA1s production cause TMD and AMKL. In euploid patients, germ-line GATA1s mutations are associated with congenital anemia, including DBA. (B) KLF1 includes an N-terminal proline-rich domain and 3 C-terminal zinc finger domains. Various missense and frameshift mutations (noted by asterisks) throughout the gene are associated with hereditary anemias, altered Lutheran blood group expression, and/or persistence of fetal hemoglobin expression. Note that E325K has a dominant negative effect. Adapted from Singleton et al.58
The first report of GATA1 mutations in defective erythropoiesis was published in 2000.14 Two half-brothers were born with severe anemia and thrombocytopenia, whereas their mother showed mild thrombocytopenia, initially thought to be caused by immune destruction. The patient's bone marrow displayed dyserythropoiesis and accumulation of dysplastic MEGs. Based on the known functions of GATA1 in these lineages and a pedigree configuration consistent with X-linked disease, mutated GATA1 was suspected and confirmed by DNA sequencing. Affected individuals harbored a mutation that substituted methionine for valine at amino acid position 205 (V205M) within the GATA1 Nf. Simultaneously, it was determined that V205 participates in the binding of GATA1 to FOG115 and that this interaction is inhibited by the V205M substitution. These findings demonstrated that the GATA1-FOG1 interaction is necessary for terminal maturation of both RBCs and MEGs, explaining the patient phenotype. Subsequently, several other groups identified different GATA1 mutations in patients with variable degrees of thrombocytopenia and anemia (Figure 3A). These mutations most commonly introduce amino acid replacements into the Nf, affecting its ability to bind FOG1, TAL1, and associated proteins (discussed below) or DNA.10,16-20 Affected patients exhibit a range of distinct and overlapping phenotypes including CDA, thrombocytopenia, erythropoietic porphyria, thalassemia, and Gray platelet syndrome. Recently, a mutation in the C terminus of GATA1 was identified in a family with X-linked Lutheran (a-b-) blood group phenotype (absent expression of the Lutheran glycoprotein) and mild macrothrombocytopenia.21 Thus, alterations in GATA1 cause a spectrum of red cell and platelet disorders whose precise characteristics relate to specific structure-function properties of the protein, many of which are not yet fully determined. Follow-up mechanistic studies have provided insights into this issue. For example, 2 missense mutations that alter the GATA1 Nf and impair DNA binding in vitro were found to have no effects on target gene occupancy when overexpressed in a cellular model for murine erythropoiesis.20 Rather, these mutations inhibited recruitment of the TAL1 complex. Understanding how different GATA1 mutations produce distinct phenotypes represents a current research challenge.
Mutations resulting in amino-truncated GATA1 (GATA1s)
Although complete loss of GATA1 leads to profound defects in the erythroid and MEG lineages (at least in mice), loss of the NAD causes less severe defects. Mutations that eliminate the NAD contribute to Down syndrome (DS)-associated transient myeloproliferative disorder (TMD) or acute megakaryoblastic leukemia (AMKL), underscoring its essential role in controlling megakaryopoiesis, particularly in the context of trisomy 21.22 In all cases, the acquired mutations lead to expression of a shortened isoform of GATA1, named GATA1s, which lacks amino acids 1-83 (Figure 3A). Loss of the N-terminal domain is associated with aberrant regulation of GATA1 target genes, defective differentiation activity, and expansion of a distinct fetal liver progenitor in vivo.22
Germ-line GATA1s-type mutations occur in individuals without DS. In these cases, the mutation does not cause TMD or AMKL but rather leads to erythroid hypoplasia. For example, a germ-line GATA1s mutation was reported in a Brazilian pedigree with defective erythropoiesis, mild defects in MEGs, and neutropenia.23 Last year, Gazda and colleagues performed exome sequencing on DBA patients lacking the typical ribosomal protein gene mutations. They discovered GATA1s mutations in 3 affected individuals from 2 unrelated pedigrees.8 A second study identified a GATA1 mutation in a different unrelated patient.24 These GATA1 mutations identify a variant form of DBA, underscore the genetic and clinical heterogeneity of this disorder, and suggest a potential mechanistic link between ribosome biogenesis and GATA1 activity.25
Mutations that disrupt GATA1 binding to DNA regulatory regions
Several mutations in DNA cis elements recognized by GATA1 have been identified. For example, the RBC Duffy antigen null phenotype, a relatively common allele in Africans that confers resistance to Plasmodium vivax malaria, is caused by a mutation that disrupts a GATA1 binding site in the Duffy antigen/chemokine receptor gene (DARC) promoter.26 X-linked sideroblastic anemia in some patients is associated with a mutation that disrupts a GATA1 binding motif in an enhancer that activates transcription of ALAS2, which encodes the enzyme catalyzing the first step in heme synthesis.27,28 Disruption of a GATA1 binding element in the promoter of the gene encoding uroporphyrinogen III synthase accounts for some cases of congenital erythropoietic porphyria.29
How do GATA1 mutations cause disease?
Many questions remain regarding GATA1-associated blood disorders. How do different GATA1 mutations confer distinct phenotypes in patients? What are the relevant target genes? How do GATA1s mutations cause anemia in children without DS, while cooperating with T21 to cause megakaryocytic defects in children with DS? Is there a relationship between GATA1 and ribosomal function in DBA?
Studies with animal models show that GATA1 is required for both RBC and MEG maturation. Gata1-null mouse embryos die in midgestation of anemia with premature cell death and/or impaired maturation of erythroid precursors. GATA1 activates and represses hundreds of genes,30-33 and its loss presumably interferes with both induction of the entire erythroid maturation program and the suppression of genes regulating stem/progenitor and or alternate lineage development. The phenotypic variability in patients with different GATA1 mutations is likely due to subtle alterations in binding to specific gene targets and/or various interacting proteins, such as FOG1, TAL1, and others,20 although the specific biochemical and structural details are largely unknown.
Interestingly, adult mice with GATA1s-type mutations do not have any obvious hematopoietic defects, although fetal hematopoiesis is perturbed,34 indicating that the manifestations of this mutation differ in humans who exhibit postnatal phenotypes. Consistent with these interspecies differences, GATA1s is produced naturally along with the full-length protein during human, but not murine, hematopoiesis. How GATA1s mutations cause anemia is not clear. One possibility is that these mutations cause reduced overall GATA1 expression (GATA1 full length + GATA1s), leading to net deficiency of the protein. In agreement, high levels of transgenic GATA1s expression can rescue anemia Gata1 gene-ablated mice.35 Alternatively, loss of the GATA1 N terminus may qualitatively alter some of its functions that are particularly important for erythroid development.
GFI1B
Growth factor independent 1 (GFI1) is a zinc finger Snail/Gfi domain (SNAG) domain-containing transcriptional repressor that controls erythroid and MEG development, in part by forming distinct complexes with GATA1 and chromatin-modifying enzymes that inhibit gene expression.36-38 Mice embryos lacking Gfi1b die in utero of anemia caused by defective erythroblast maturation and they also harbor developmentally arrested MEGs,39 similar to the phenotype caused by loss of GATA1. GFI1B regulates proliferation and differentiation of MEPs through effects on transforming growth factor-β signaling.40 One study found a heterozygous frameshift mutation in the GFI1B gene of a family with an autosomal dominant bleeding disorder, macrothrombocytopenia, and platelet dysfunction.41 Affected individuals also exhibited mild RBC abnormalities including anisopoikilocytosis and increased red cell distribution width. In addition, a GFI1B mutation causing the production of a dominant interfering protein was recently described to cause Gray platelet syndrome.42
KLF1
KLF1 is the founding member of a family of Sp1 transcription factor (Sp1)-related TFs that contains 3 C-terminal C2H2 zinc finger DNA binding domains, as well as an N-terminal proline-rich region (Figure 3B).43,44 Klf1-null embryos die in midgestation from anemia with a β-thalassemia-like syndrome.45,46 KLF1 activates expression of key erythroid genes including those that encode β-globin, heme biosynthetic enzymes, RBC membrane proteins, and cell cycle regulators.47-49 Chromatin immunoprecipitation coupled with next-generation sequencing detected KLF1 binding at the regulatory regions of numerous erythroid genes, frequently in close proximity to GATA1-bound sites.44 Another study found that GATA1, KLF1, and TAL1 co-occupy many erythroid-expressed genes.50 KLF1 undergoes numerous post-translational modifications, including phosphorylation, sumolyation, acetylation, and ubiquitinylation. These changes are believed to regulate its interactions with partner proteins and to control its activity in vivo.50 In MEPs, KLF1 expression contributes to the specification of erythroid cell fate by competing with the MEG factor FLI1.1,2
Mutations affecting KLF1 function cause isolated anemia without thrombocytopenia. Mutations that disrupt KLF1 binding to a cognate element in the human β globin gene (HBB) promoter cause β+ thalassemia.9 Mutations in the human KLF1 gene itself were first discovered in individuals with the Lutheran (a-b-) blood group phenotype,51 which also occurs in GATA1-mutant patients (discussed previously and reviewed in ref. 52). Twenty-one of 24 Lutheran (a-b-) blood group individuals studied harbored 9 different heterozygous KLF1 loss-of-function mutations (Figure 3B). These individuals exhibited no anemia or overt RBC abnormalities. Therefore, 1 allele of KLF1 appears to suffice for normal erythro-poiesis, similar to what occurs in mice. Presumably, the gene encoding the Lutheran glycoprotein is positively regulated by GATA1 and KLF1 and is highly sensitive to dosage of the latter.
Nan (neonatal anemia) mice harbor a Klf1 missense mutation resulting in an E339D substitution within the second zinc finger of the protein.53,54 Homozygous embryos die in midgestation of anemia, whereas heterozygous animals display severe hemolytic anemia resembling hereditary spherocytosis.55 Nan heterozygous mutant RBCs showed reduced expression of membrane cytoskeleton proteins including band3 and proteins 4.1 and 4.2, which are commonly deficient in hereditary spherocytosis.53 In contrast to loss of a single allele of Klf1, which produces minimal effects on erythropoiesis, the Nan mutation acts dominantly, suggesting that the E339D substitution somehow interferes with the functions of the coexpressed wild-type KLF1 protein.
Analogous to the Nan mouse, a heterozygous human KLF1 mutation at E325 (within the second zinc finger of the KLF protein) is associated with CDA56 (Figure 3B). Affected individuals display severe anemia, jaundice, and high levels of circulating nucleated RBCs. Interestingly, KLF1 E325K patients also express high levels of fetal hemoglobin (HbF, α2γ2), suggesting a role for KLF1 in the γ to β globin switch that occurs perinatally (discussed in the next section). Of note, a recent study demonstrated that KLF1 mutations cause severe congenital hemolytic anemia associated with a deficiency of pyruvate kinase in red cells and persistence of embryonic globin synthesis.57 Affected individuals also displayed the Lutheran (a-b-) blood group phenotype.
How do KLF1 mutations cause disease?
Structure function assays examining various KLF1 mutations have provided interesting insights. For example, expression of the E339D mutant was found to interfere with the ability of KLF1 to bind and regulate a subset of targets depending on 1 specific nucleotide residing in the middle of the KLF1 consensus DNA binding motif.53 Binding to genes with the consensus containing a cytidine, such as Ahsp, was maintained by E339D, whereas binding to genes with the consensus containing thymidine, including Hbb and E2f2, was disrupted by the mutation. Similar to murine E339D, the human E325K mutation produces a dominant-negative effect on KLF1-mediated transcription, including failure to bind and properly regulate a subset of target genes including CD44 and AQP1.56 Another report compared the DNA binding of mutations associated with the Lutheran (a-b-) blood group phenotype to ones seen in CDA or mild anemia.58 This study found that mutants associated with the Lutheran (a-b-) phenotype (eg, R328H; Figure 3B) failed to bind DNA, resulting in an overall reduction in the level of functional KLF1 and a mild phenotype. A mutant associated with mild anemia (K332Q) showed reduced binding to a number of DNA targets, but near normal binding to Hbb. Finally, the CDA E325K mutant displayed reduced binding to all promoters examined. In this case, however, the mutant evokes a strong phenotype even in the presence of the wild-type allele. This effect may reflect the ability of the mutant to destabilize TF complexes.58
KLF1/BCL11A regulatory axis in hemoglobin switching
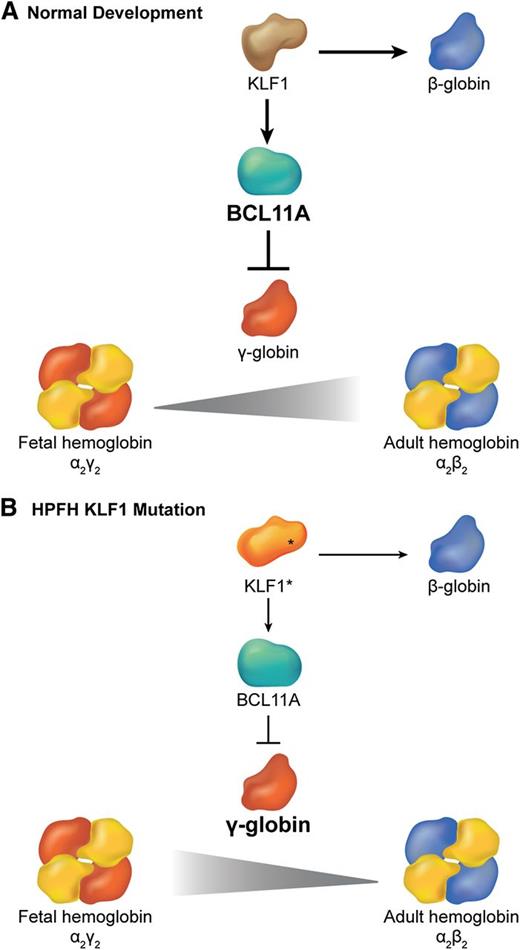
Different β globin-like genes are expressed sequentially during ontogeny.59 γ globin is expressed predominantly during most of human fetal gestation to generate HbF (α2γ2). Shortly after birth, there is a gradual switch from γ to β globin gene transcription with the resultant production of adult hemoglobin (α2β2). Haploinsufficiency of KLF1 and the E325K mutation are associated with hereditary persistence of fetal hemoglobin (HPFH),60 a condition characterized by continued high-level expression of HbF in adult RBCs. Given that increased levels of HbF alleviate common hemoglobinopathies such as β-thalassemia and sickle cell anemia, understanding and manipulating the γ (HBG) to β (HBB) globin gene switch is of major clinical importance. Borg et al used genetic linkage analysis to identify chromosomal regions that are associated with HPFH in a Maltese family.60 They discovered a heterozygous K288X KLF1 mutation that is predicted to disrupt the DNA binding zinc finger domain (Figure 3B). Transcriptional profiling in patient erythroblasts demonstrated downregulation of many mRNAs, including HBB (a known KLF1 target) and B-cell lymphoma 11A (BCL11A), which encodes a TF that represses γ globin after birth.61-65 Subsequent chromatin immunoprecipitation studies showed that wild-type KLF1 binds BCL11A promoter elements in adult, but not fetal liver erythroid progenitors.60 Thus, KLF1 appears to mediate γ to β globin switching by binding the BCL11A gene and activating its transcription. Consistent with this model, deletion of an upstream Klf1 enhancer in mice harboring a human β globin locus bacterial artificial chromosome inhibited expression of the endogenous murine Klf1 and Bcl11a genes and increased the human γ-globin/β-globin ratios in erythroblasts.66 These genetic findings indicate that alterations in a KLF1-BCL11A transcription axis (Figure 4) account for some human HPFH. Several studies suggest that inhibition of human BCL11A (through KLF1-dependent or -independent mechanisms) can de-repress γ globin transcription and thereby alleviate the clinical manifestations of some human hemoglobinopathies, including sickle cell anemia and β-thalassemia.67,68
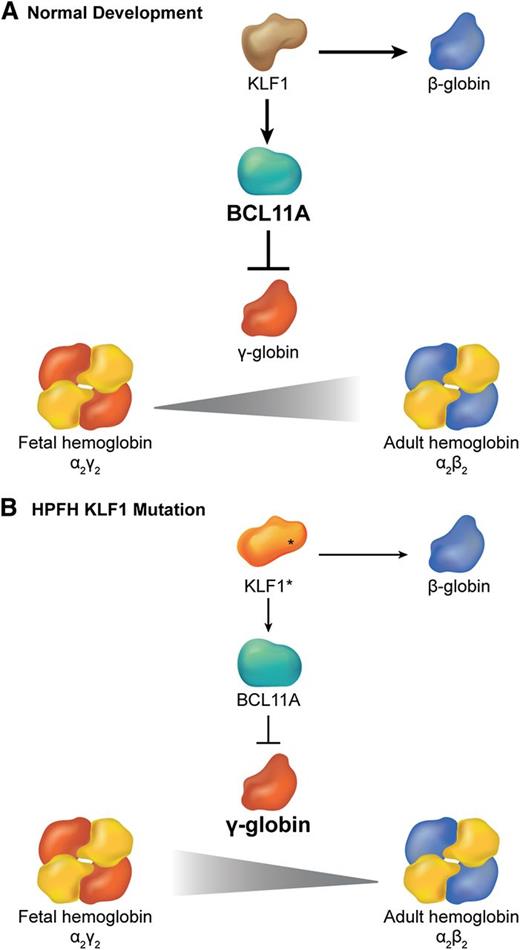
Interplay between KLF1 and BCL11a in the regulation of hemoglobin switching. (A) During normal development, KLF1 activates transcription of the BCL11A gene, which in turn represses γ globin expression promoting the switch from fetal (α2γ2) to adult (α2β2) hemoglobin. Simultaneously, high levels of KLF1 activate β-globin gene expression. (B) In some cases of HPFH, haplo-insufficiency of KLF1 (KLF1*) is associated with reduced BCL11A expression, which allows for persistence of fetal hemoglobin. Simultaneously, reduced KLF1 fails to activate β-globin expression.
Interplay between KLF1 and BCL11a in the regulation of hemoglobin switching. (A) During normal development, KLF1 activates transcription of the BCL11A gene, which in turn represses γ globin expression promoting the switch from fetal (α2γ2) to adult (α2β2) hemoglobin. Simultaneously, high levels of KLF1 activate β-globin gene expression. (B) In some cases of HPFH, haplo-insufficiency of KLF1 (KLF1*) is associated with reduced BCL11A expression, which allows for persistence of fetal hemoglobin. Simultaneously, reduced KLF1 fails to activate β-globin expression.
Predicting new human hematopoietic disease alleles through genetic and biochemical studies
GATA1, KLF1, and Gfi1b, which are disrupted in human blood disorders (see previous sections), bind several other hematopoietic TFs to activate and repress gene expression during erythro-megakaryopoiesis. Mutations in genes encoding any members of these complexes could potentially result in anemia and/or thrombocytopenia. Salient candidate genes include the following:
TAL1 (previously also called stem cell leukemia [SCL]) encodes a basic helix-loop-helix TF that participates in the formation of hematopoietic stem cells during murine embryogenesis and erythro-megakaryopoiesis postnatally.69-72 TAL1 regulates gene expression via formation of a multisubunit complex that includes E2A, LIM binding domain 1 (LDB1), LIM domain only 2 (LMO2), and GATA proteins.73 Somatic gene rearrangements resulting in TAL1 overexpression are associated with a subset of childhood T-cell acute lymphoblastic leukemias.74 Given the essential role for TAL1 in erythropoiesis in mice,69 it is possible that inherited hypomorphic or neomorphic mutations could cause CDA and/or thrombocytopenia. Such mutations may be associated with pleomorphic phenotypes because TAL1 is expressed in numerous nonhematopoietic tissues including endothelial and nerve cells.
GATA binding protein 2 (GATA2) encodes a GATA1-related protein with essential roles in normal and malignant hematopoiesis.75,76 Gata2 knockout embryos die of anemia with defective production of hematopoietic stem and progenitor cells.77,78 Germ-line heterozygous GATA2 loss-of-function mutations cause several related diseases associated with myelodysplasia and/or immunodeficiency, including Mono-Mac syndrome, Emburger syndrome, and familial predisposition to acute myeloid leukemia.79-81 The GATA2 gene is expressed in early RBC precursors and downregulated during subsequent phases of maturation, in part through direct repression by GATA1. This “GATA switch,” whereby GATA1 replaces GATA2 at various sites in the genome, appears to be essential for normal erythropoiesis.82-84 Thus, cis elements in the GATA2 gene that selectively mediate its downregulation during erythropoiesis represent potential target regions for mutations that cause CDA syndromes.85
FOG1 encodes a multi-zinc finger nuclear protein that does not bind DNA on its own, but provides essential functions in RBC and MEG development via physical interactions with the GATA1 Nf.13,86 Fog1−/− mice die in midgestation with impaired erythroid progenitor and MEG maturation, phenocopying loss of GATA1.87 GATA1 Nf missense mutations that impair binding to FOG1 cause CDA and/or thrombocytopenia, emphasizing the importance of this association.20 Based on these findings, germ-line FOG1 mutations may cause anemia and/or thrombocytopenia. Such mutations could also be associated with congenital heart disease, as indicated by conditional gene targeting studies of Fog1 in mice.88
LDB1 and LMO2 encode nuclear adaptor proteins that form complexes with GATA1, GATA2, and TAL1 in numerous hematopoietic cell types including stem/progenitor, erythroid, and MEGs.89 These adaptors colocalize with GATA1, KLF1, and TAL1 at numerous RBC- and MEG-expressed genes. LDB1 self-oligomerizes and this property mediates long-range promoter-enhancer DNA looping interactions that activate or repress gene expression. Consistent with these findings, ablation of the Ldb1 or Lmo2 genes in mice cause severe anemia. Germ-line mutations in the corresponding human genes could cause inherited anemia, thrombocytopenia, or other blood abnormalities. Such mutations may cause additional developmental abnormalities, because LDB1 and LMO2 appear to have essential functions in many tissues.
The NFE2 gene encodes nuclear factor-erythroid 2, discovered as protein heterodimer that binds conserved functional elements present in globins and many other erythroid and megakaryocytic genes.90,91 The NF-E2 complex is comprised of a hematopoietic specific 45-kDa basic leucine zipper protein and a widely expressed 18-kDa subunit of the Maf family. The NFE2 gene is up-regulated in polycythemia vera where it is believed to promote erythropoietin-independent erythropoiesis and expansion of the hematopoietic stem cell and common myeloid lineages.92,93 Mice engineered to overexpress NF-E2 protein develop similar phenotypes.94 Moreover, patients with myeloproliferative neoplasms harbor somatic NFE2 mutations that may activate protein function.95 Thus, excessive NF-E2 activity appears to drive abnormal proliferation of erythroid, myeloid, and MEG lineages.
Nfe2−/− mice exhibit profound thrombocytopenia with minimal anemia, most likely due to the presence of similar factors in erythroid cells with redundant functions.96-98 Thus, murine studies predict that human NFE2 inactivating mutations would cause thrombocytopenia but not anemia. However, it is important to note that murine mutagenesis studies do not always predict human phenotypes. A germane example is SEC23B, which encodes a member of the coat protein complex II that facilitates exit of secreted proteins from the endoplasmic reticulum. Mutations in SEC23B cause CDA type II,99 whereas ablation of the same gene in mice causes pancreatic secretory dysfunction but no anemia.100 These disparate phenotypes may be caused by species-specific shifts in the functions of SEC23B and its closely related paralog SEC23A.
Quantitative trait loci-hematopoietic TF interactions
As discussed earlier in this review, mutations in TF binding DNA cis elements residing within promoters and enhancers can cause human anemias. Interestingly, mutations of this type can also exert subtle effects that are nonetheless clinically significant. Many quantitative trait loci identified by genome-wide association studies map to proximal cis elements that regulate the expression of nearby (or distant) genes,101 a concept that is relevant to RBC disorders. For example, genome-wide association studies have identified numerous loci that regulate RBC traits, including persistence of fetal hemoglobin expression in adults.102,103 Of particular interest, a DNA sequence variant that inhibits binding of GATA1 and TAL1 to a BCL11A gene enhancer causes elevated fetal hemoglobin in adult RBCs.104 This finding is of major clinical significance because disruption of this enhancer region by genome editing105 represents a potential means to de-repress γ globin synthesis for treating common hemoglobinopathies including sickle cell anemia and β thalassemia. Another interesting quantitative trait locus residing within the cyclin D3 (CCND3) gene regulates human RBC size and number. In this case, a nucleotide substitution modifies a TAL1 binding site within an erythroid-specific enhancer to alter CCND3 expression, thereby impacting cell division and cell size during erythroid maturation.106 These recent discoveries of cis element mutations/polymorphisms that are associated with human RBC traits and anemias (discussed earlier and in Figure 2E) likely represent the tip of the iceberg. Genetic alterations in cis elements that regulate TF binding probably represent a relatively common etiology for blood diseases and an even more widespread mechanism for variation of RBC traits within populations. Of course, these concepts are generalizable to virtually all medically relevant quantitative traits and heritable disorders. Importantly, most cis element mutations would not be ascertained through whole genome exome sequencing.
Conclusions
Mutations in genes encoding hematopoietic TFs or their cognate cis elements are becoming increasingly appreciated as causes for congenital anemias and/or thrombocytopenias. Analysis of these mutations has taught us a great deal about TF function and mechanisms of gene expression. Two scientific advances have facilitated the recognition of these mutations: First, well-characterized phenotypes arising from murine loss-of-function studies (ie, gene knockouts) identify candidate TF genes for targeted examination in unexplained inherited blood disorders. Second, nonbiased genome sequencing of affected human patients and pedigrees is increasingly used for genetic diagnosis.107 Both technologies are rapidly evolving. For example, new genome editing methods, such as transcription activator-like effector nucleases and the clustered regulatory interspaced short palindromic repeats/Cas system, facilitate gene targeting in mice and human pluripotent stem cells,108 which should increase the repertoire of candidate TF mutations predicted to affect human hematopoiesis. Second, global exome and whole genome sequencing are becoming more available and cost effective for diagnostics. The latter, in particular, permits identification of cis element mutations outside of protein coding regions. Moreover, functional characterization of human and murine genomes, largely by The Encyclopedia of DNA Elements Consortium, is identifying thousands of potential tissue-specific enhancers that represent candidate regions for disease-associated mutations.109-111 Together, these technologies should identify causal mutations for most Mendelian disorders and many multigenic ones in the near future, including those that affect hematopoiesis. Understanding precisely how these mutations regulate TF function and gene expression represents an exciting and formidable long-term challenge.
Acknowledgments
The authors thank Gerd Blobel, Amy Campbell, and Vijay Sankaran for reviewing this manuscript.
Work on erythropoiesis in M.J.W.'s laboratory is supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK092318, P30DK090969, and R01DK065806, US Department of Defense grant BM090168, US Israel Binational Science Foundation grant 2009239, and the Jane Fishman-Grinberg Endowed Chair for Stem Cell Research. Research on erythropoiesis in J.D.C.’s laboratory is supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant DK101329 and the Samuel Waxman Cancer Research Foundation.
Authorship
Contribution: J.D.C. and M.J.W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John D. Crispino, Northwestern University, Division of Hematology/Oncology, 303 East Superior St, Lurie Bldg, Room 5-113, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu; and Mitchell Weiss, Children’s Hospital of Philadelphia, Division of Hematology, Room 316B ARC, 3615 Civic Center Blvd, Philadelphia PA 19104; e-mail: weissmi@email.chop.edu.