In this issue of Blood, Chan et al report on an innovative study using molecular dynamics and mapping to identify and produce a promising bioengineered clone of Escherichia coli asparaginase (ASNase), mutated at Q59L position (Gln59Leu). The Q59L variant ASNase has undetectable glutaminase coactivity, while retaining its ASNase activity.1
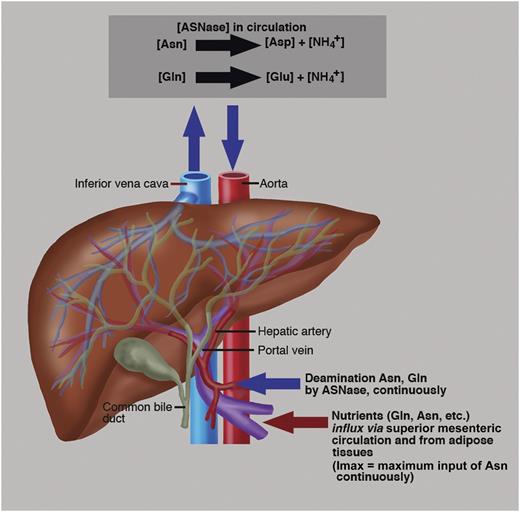
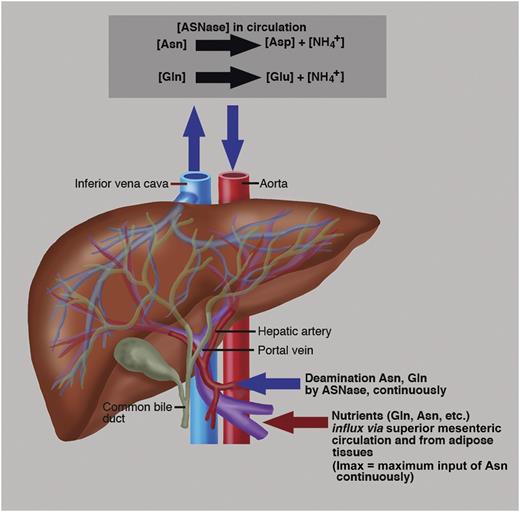
ASNase in circulation continuously deaminates Asn with a Km = 29 μM and to a lesser extent Gln with a Km ∼900 μM.2 The hepatic portal vein is the blood vessel that influxes blood via the mesenteric circulation from the gastrointestinal tract and spleen to the liver. This blood is rich in nutrients that have been extracted from food, like amino acids, Asn, Gln, etc.2 The liver processes these nutrients; adipose tissues also provide Gln.10 The liver also synthesizes Asn de novo from Gln aspartate and ATP, a reaction catalyzed via ASNS.2 The sum of the Asn sources is described biochemically as maximum input (Imax) into circulation. Therefore, an effective ASNase must deplete not only Asn but also Gln to reduce the ASNS contribution of Asn biosynthesis.2 Most importantly, in rapidly proliferating malignant cells, Gln and Asp are used as precursors of -N-C- links in the de novo nucleoside biosynthesis. Therefore, depletion of Gln by an ASNase with glutaminase coactivity contributes to the inhibition of protein and DNA biosynthesis. Professional illustration by Marie Dauenheimer.
ASNase in circulation continuously deaminates Asn with a Km = 29 μM and to a lesser extent Gln with a Km ∼900 μM.2 The hepatic portal vein is the blood vessel that influxes blood via the mesenteric circulation from the gastrointestinal tract and spleen to the liver. This blood is rich in nutrients that have been extracted from food, like amino acids, Asn, Gln, etc.2 The liver processes these nutrients; adipose tissues also provide Gln.10 The liver also synthesizes Asn de novo from Gln aspartate and ATP, a reaction catalyzed via ASNS.2 The sum of the Asn sources is described biochemically as maximum input (Imax) into circulation. Therefore, an effective ASNase must deplete not only Asn but also Gln to reduce the ASNS contribution of Asn biosynthesis.2 Most importantly, in rapidly proliferating malignant cells, Gln and Asp are used as precursors of -N-C- links in the de novo nucleoside biosynthesis. Therefore, depletion of Gln by an ASNase with glutaminase coactivity contributes to the inhibition of protein and DNA biosynthesis. Professional illustration by Marie Dauenheimer.
ASNases are integral components of childhood acute lymphoblastic leukemia (ALL) regimens, which act to deaminate asparagine (Asn) and glutamine (Gln). Bacterial ASNase proteins have different biochemical and pharmacological properties with different side effects, predominantly due to the variable glutaminase coactivity in addition to the ASNase activity. Understanding the properties of various l-ASNases would allow a better understanding of their catalytic and therapeutic features, thus enabling more accurate predictions of therapeutic effects.2
To understand the activity of ASNase, one must comprehend its enzymatic actions. Four independent catalytic sites are located at the intersubunit interface of the intimate dimers.2,3 Enzymatic activity depends on the classic 2-step nucleophilic attack mechanism in E coli ASNase, initiated by WAT1355 and the cooperation of the highly conserved amino acids of Thr12-Tyr25-Glu283 and Ser58-Gln59-Thr89-Asp90-Lys162 from subunit A, and Asn248C and Glu283C from subunit C, respectively, which are interconnected by strong hydrogen bonds.4,5 The Thr12-Lys162-Asp90 group acts to deprotonate a water molecule (WAT1355) which subsequently binds to the substrate. The second triplet, Thr12-Ty25-Glu283, stabilizes the resulting tetrahedral intermediate.3 ASNase crystallography data suggest that a peptide loop (amino acid residues 14-27) is highly flexible and shields the active site during catalysis in the enzyme.2,5,6 Apparently, the length of the flexible lid loop varies among bacterial ASNases depending on their ability to accommodate Gln as a substrate.2
Gln59 resides adjacent to Asp90, and >120° in opposite orientation to the nucleophile; hence, it does not take part in the deamination reaction.5 However, this article strongly suggests that its replacement (Q59L) excludes Gln from the catalytic site. Observation of the ASNase active site supports that the Q59L form should reduce glutaminase activity with impaired substrate binding and stabilization of the transition state. Chan et al have clearly demonstrated this biochemical effect.1
The authors next explored the antileukemia activity of this Q59L protein, demonstrating it to be “potentially active” against Asn synthetase (ASNS)–negative cells, but inactive against cells expressing measurable ASNS. The authors further propose that in ASNS-negative ALL cells, the Q59L variant ASNase protein may be less toxic to patients, while providing a large therapeutic index.
One of the limitations of the study is the absence of consideration of the many modes of ASNase resistance in patients. The most common mechanism of ASNase resistance is due to antibody (Ab) formation.2 Upon initiation of combination chemotherapy, there is a massive release of cellular debris from dying ALL cells. ALL lysosomal endopeptidases exist in the cellular material and can cleave ASNase in the Asn24 position, thus exposing the pentapeptide amino acids 24 to 29.2,7 This peptide is one of the most antigenic peptides and initiates the host immune response to the bacterial protein(s) causing the formation of a neutralizing Ab at the active site.2-4 It is noteworthy that the authors have not established whether the variant ASNase protein is cross-reactive with anti-E coli ASNase Abs.
ALL cells which manage to survive the initial onslaught of combination chemotherapy, whether due to Ab formation or other mechanism, undergo a variety of adaptations which contribute to their potential to cause relapse.2 Under severe Asn and to a lesser extent Gln depletion, ASNS is upregulated intracellularly. Thus, even ALL cells which initially do not express significant levels of ASNS could become ASNS expressing. Moreover, a wealth of research illustrates that this is associated with cMyc and Bcl-2 upregulation. Fine et al found that more than 800 genes alter their expression after exposure to ASNase.8 Under Asn and Gln deficiencies, cells activate the general control nonderepressible 2 (GCN2) kinase, which phosphorylates the α-subunit of the eukaryotic initiation factor 2 (eIF2α).9 This step represents a fundamental metabolic switch, causing a shift in protein biosynthesis from the high-energy demanding 5′-cap messenger RNA (mRNA) entry into the cellular translational machinery to the internal mRNA entry site, which is less energy dependent.9
Another important metabolic regulatory switch affected by ASNase activity is the mammalian target of rapamycin (mTOR) pathway; however, mTOR signaling is sensitive to Gln levels.3 Therefore, depletion of Gln is of paramount importance to eliminate refractory leukemia cells. Moreover, the activity of Gln synthetase (GS) in adipose cells is increased upon ASNase treatment.10 Furthermore, there is evidence that ASNS expression in cells of the ALL microenvironment can contribute to their ability to provide Asn to nearby ALL cells.2,10 Thus, even ALL cells which do not express ASNS may be protected from Asn depletion by production in neighboring cells, using Gln as a source2,10 (see figure). The upregulation of ASNS becomes apparent in high risk and early relapsed patients. In order to benefit these patients, intense pegaspargase regimens or Erwinase-containing regimens are used,2,10 which obtain durable second complete remissions. These intense ASNase dosing regimens result in higher enzymatic trough levels, which result in the pharmacodynamic benefit of deamination of Asn and depletion of Gln to a significant degree, nullifying the prosurvival effects of GS and ASNS.2 It should be emphasized that Gln and Asn are precursors of the de novo biosynthesis of pyrimidines and purines for DNA replication. Glutamine depletion, therefore, could be significant not only for inhibition of protein synthesis, but also for inhibition of DNA synthesis.
I think that this bioengineered ASNase protein may find considerable use in the treatment of standard-risk children (low blast count, good status, etc) diagnosed early in their disease. However, the unmet needs in the treatment of refractory leukemia include bioengineered ASNase proteins that are resistant to proteolytic hydrolysis by lysosomal enzymes2 and a bioengineered protein with various changes with highly conserved amino acids from the Enterobacteriaceae family with optimal ASNase and glutaminase activity. Ideally, these ASNases should be built on an E coli backbone for greater physiological thermal stability and optimize the glutaminase coactivity levels.
Conflict-of-interest disclosure: The author declares no competing financial interests.