Key Points
Trisomy 12 CLL cells exhibit upregulated integrin signaling and enhanced VLA-4-directed adhesion and motility.
The increased expression of β2-integrins on trisomy 12 CLL cells is modulated by intercurrent NOTCH1 mutations.
Abstract
The leukocyte adhesion cascade is important in chronic lymphocytic leukemia (CLL), as it controls migration of malignant cells into the pro-survival lymph node microenvironment. Circulating trisomy 12 CLL cells have increased expression of the integrins CD11a and CD49d, as well as CD38, but the tissue expression of these and other molecules, and the functional and clinical sequelae of these changes have not been described. Here, we demonstrate that circulating trisomy 12 CLL cells also have increased expression of the integrins CD11b, CD18, CD29, and ITGB7, and the adhesion molecule CD323. Notably, there was reduced expression of CD11a, CD11b, and CD18 in trisomy 12 cases with NOTCH1 mutations compared with wild type. Trisomy 12 cells also exhibit upregulation of intracellular integrin signaling molecules CALDAG-GEFI, RAP1B, and Ras-related protein ligand, resulting in enhanced very late antigen-4 [VLA-4] directed adhesion and motility. CD38 expression in CLL has prognostic significance, but the increased CD38 expression in trisomy 12 CLL cells must be taken into account in this subgroup, and the threshold of CD38 positivity should be raised to 40% for this marker to retain its prognostic value. In conclusion, trisomy 12 CLL cells exhibit functional upregulation of integrin signaling, with β2-integrin expression being modulated by NOTCH1 mutation status.
Introduction
Chronic lymphocytic leukemia (CLL) is a disease of considerable clinical and genetic heterogeneity. Trisomy 12 is the third most common cytogenetic abnormality and has several distinguishing features including abnormal morphology and increased prevalence of NOTCH1 mutations.1,2 Although trisomy 12 is present in approximately 16% of cases of CLL, the prevalence of this cytogenetic abnormality is significantly higher in small lymphocytic lymphoma (SLL) where it is present in 28% of cases.3 Furthermore, acquisition of trisomy 12 also has been recently implicated in a third of cases of Richter’s transformation.4
The increased prevalence of trisomy 12 in these lymphomas is of particular interest in light of studies reporting increased expression of the α-integrins CD11a and CD49d on trisomy 12 CLL cells.5,6 The heterodimeric integrins CD11a/CD18 (LFA-1), CD11b/CD18 (Mac-1), CD49d/CD29 (very late antigen-4 [VLA-4]), and CD49d/ITGB7 are cell surface transmembrane proteins involved in the inducible adhesion of leukocytes to vascular walls during the process of transendothelial migration from the bloodstream into the tissues. This process is particularly important in CLL as it allows the malignant cells to enter lymphoid organs where they receive growth and survival signals and are protected from chemotherapy by a network of interactions with the lymph node (LN) microenvironment.7 Despite previous reports regarding CD11a and CD49d, a full characterization of molecules involved in leukocyte transmigration including other integrins, selectins, and adhesion molecules has not been described. Furthermore, studies examining the relative expression of integrins in the LNs, the degree of activation of integrin signaling pathways, and the functional impact of changes in integrin expression are lacking.
In this report, we demonstrate that circulating trisomy 12 CLL cells have increased expression of the integrins CD11b, CD18, CD29, and ITGB7, and the adhesion molecule CD323, in addition to increased expression of CD11a and CD49d. Notably, these changes are modulated by NOTCH1 mutation status, with NOTCH1 mutated trisomy 12 cases having lower expression of CD11a, CD11b, and CD18 compared with wild-type. Trisomy 12 CLL cells also have upregulation of integrin signaling pathways resulting in increased ligand binding and enhanced VLA-4-directed adhesion and motility. Finally, we also demonstrate that the increased expression of CD38 on trisomy 12 CLL cells means that CD38 cannot be used as a surrogate marker of IGVH gene mutation status in this subgroup. Furthermore, the threshold of CD38 positivity should be raised to 40% in the presence of trisomy 12 for this marker to retain its prognostic value.
Material and methods
Patients
Peripheral blood (PB) samples were obtained from 118 untreated CLL patients from the tissue core maintained by the CLL Research Consortium (CRC) according to the guidelines established by the Health Insurance Probability and Accountability Act. Further PB samples were obtained for a separate cohort of 15 trisomy 12 CLL patients with known NOTCH1 mutation status from the CRC tissue core.1 Data from the CRC database for a cohort of 463 patients with trisomy 12 detectable by fluorescence in-situ hybridization was used for the CD38 analysis. Tissue cores from LN biopsies were obtained from 31 CLL patients and 27 healthy controls from the tissue bank maintained by the Department of Haemato-Oncology of St. Bartholomew’s Hospital, London, UK. PB samples were also obtained from a control group of 25 age-matched healthy volunteers with a median age of 64 years (range, 49-72 years). All patients had consented for sample storage in accordance with the Declaration of Helsinki, and all studies were approved by the institutional review board.
Monoclonal antibodies
The following directly conjugated monoclonal antibodies were used in this study: CD5-PerCPCy5.5, CD11a-FITC, CD19-APC-eFluor780, CD29-APC, CD31-PECy7, CD38-PECy7, CD49d-PE, CD99-FITC, CD102-FITC, CD162-APC, CD323-PE, and ITGB7-FITC, and were all obtained from eBioscience. CD11b-APC, CD18-APC CD62L-PE, and CD321-PE were all obtained from BD Biosciences. For immunohistochemistry, primary antibodies specific for CD79a, CD18, and ITGB7 were from Sigma Prestige; anti-CD11a was from R&D; anti-CD29 was from Fisher Scientific; and anti-Ki67 was obtained from Dako. Further details of all monoclonal antibodies used are provided in supplemental Table 1, available on the Blood Web site.
Isolation of PBMCs and lymphocyte subsets
PB samples were diluted 1:1 with phosphate buffered saline (PBS) prior to separation of PB mononuclear cells (PBMCs) by density gradient centrifugation. Cells were washed in RPMI 1640 supplemented with 10% fetal calf serum (PAA Laboratories) and 25 mg gentamicin (Gibco). Where necessary, CD19+ healthy B cells or CLL cells were positively selected using CD19+ microbeads (Miltenyi Biotec).
Immunofluorescence staining and flow cytometric analysis
For surface staining, PBMCs were washed twice in PBS containing 2% fetal calf serum (staining buffer). Cells were then incubated with directly conjugated monoclonal antibodies for 30 minutes at 4°C. The cells were then washed and resuspended in staining buffer with 250 ng/mL 4,6 diamidino-2-phenylindole (DAPI; Invitrogen), and kept at 4°C until analysis. Flow cytometry was performed on a BD Fortessa flow cytometer with subsequent analysis using FlowJo software (Tree Star). Analysis was performed after gating on live singlet cells.
Immunohistochemistry
Tissue microarrays of triplicate 1-mm diameter cores were prepared from paraffin blocks using a manual tissue arrayer (Beecher Scientific) as previously described.8 “CLL-cell rich” cores with >80% of cells positive for CD79a were used for analysis. A panel of monoclonal antibodies specific for CD11a, CD18, CD29, ITGB7, and Ki67 was used to determine integrin expression and proliferation. The primary antibody reaction was detected using a peroxidase-labeled detection system (Super Sensitive Polymer-HRP IHC Detection System; BioGenex). The slides were scanned with an Olympus BX61 microscope. Immunostaining was quantified by computerized image analysis using the DensitoQuant tool in Pannoramic Viewer (3DHistTech). Further details are provided in the supplemental materials and in “Material and methods.”
Quantitative RT-PCR
RNA was extracted from CLL cells or healthy B cells using the RNeasy Plus Mini Kit (Qiagen) and converted to complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit according to manufacturer’s instructions. The cDNA was subsequently used in 20 μL quantitative real time polymerase chain reaction (RT-PCR) reactions using Applied Biosystems Taqman Gene Expression Assays. Reactions were performed in duplicate on Applied Biosystems 7900HT Fast RT-PCR machine using the standard thermal cycler protocol. The expression of each target gene was calculated relative to the endogenous control gene (TATA-binding protein) using the 2−ΔΔCT method. All of the primer/probe sets (RASGRP2, RAP1B, RASSF5, RAP1A, PXN, TLN1, and VCL) and reaction materials were purchased from Applied Biosystems.
Soluble ICAM-1 and VCAM-1 binding assay
Frozen CLL cells or healthy B cells were thawed in full medium and rested overnight at 37°C; 5% CO2. They were then washed in Hank’s Balanced Salt Solution (HBSS) containing 1mM CaCl2 and MgCl2 (Invitrogen) with 20mM HEPES (Invitrogen)(Binding buffer) at 37°C. Cells were exposed to 3mM MnCl2(Sigma) or 50ng/mL PMA in the presence of ICAM-1/Fc (20 μg/mL; R&D Systems) or VCAM-1/Fc (20 μg/mL; R&D Systems) or an equal volume of binding buffer (control) for 3 minutes at 37°C. Cells were then immediately fixed on ice in HBSS with 1% paraformaldehyde and washed in binding buffer before being labeled with PE-conjugated anti-human IgG Fc antibody (Biolegend) for 30 minutes at 4°C. ICAM-1 or VCAM-1 binding was measured using flow cytometric measurement of PE median fluorescence intensity.
Static ICAM-1/VCAM-1 adhesion and LFA-1/VLA-4-directed motility assays
Hi-Q4 culture dishes (Nikon) plates were coated overnight at 4°C with 3 μg/mL intercellular adhesion molecule (ICAM)-1-Fc or 3 μg/mL vascular cell adhesion molecule (VCAM)-1-Fc and blocked with 2% bovine serum albumin in PBS at room temperature for 1 hour.9 Dishes were washed twice and 3.5 × 105 lymphocytes in 350 μL of binding buffer with 1 μg/mL CXCL12 (R&D) added. MnCl2 was not used for these assays. Images were taken with a Nikon BioStation IM microscope (Nikon UK Ltd, UK), using a 20× objective lens and the BioStation software (Nikon) at 30-second intervals for 1 hour. For the adhesion assay, the proportion of cells with a spread “adherent” conformation was analyzed after 30 minutes stimulation; a minimum of 100 cells were counted. To calculate cell motility, the cells were tracked and analyzed with NIS-Element AR software (Nikon) and the average velocity (μm/second) of at least 50 cells analyzed.
Statistical analysis
All data sets were subject to normality testing using the Shapiro-Wilk normality test. An unpaired Student t test was used for the analysis of differences between the groups for all data sets could be accurately modeled by a Gaussian distribution; this did not apply to the 2-sided Mann-Whitney U test was used. For comparison of 3 groups, the Kruskal-Wallis test was used with Dunn’s post-test for multiple comparisons. Comparison of Kaplan-Meier survival curves was performed using the log rank (Mantel-Cox) test. P < .05 values were considered statistically significant. Recursive partitioning (using the RPART macro in the R programming language) was performed using dividing rules based on the likelihood ratio test to examine the optimal split of CD38, which dichotomize the patients into groups that maximally discriminates between treated/untreated patients.
Results
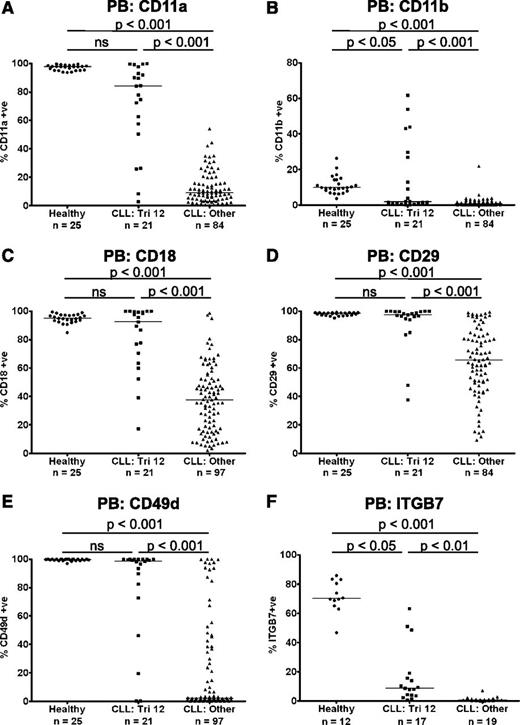
Preserved expression of the integrins CD11a, CD11b, CD18, CD29, CD49d, and ITGB7 on trisomy 12 CLL cells
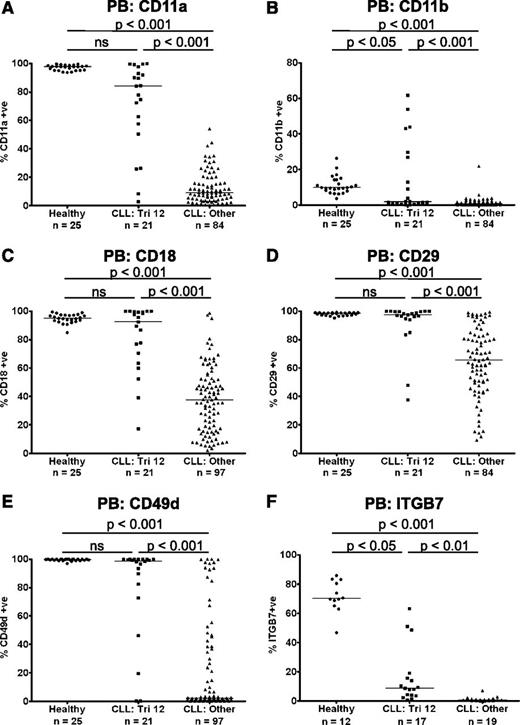
The heterodimeric integrins CD11a/CD18 (LFA-1), CD11b/CD18 (Mac-1), CD49d/CD29 (VLA-4), and CD49d/ITGB7 are cell surface transmembrane proteins involved in the inducible adhesion of leukocytes to vascular walls during the process of transendothelial migration from the bloodstream into the tissues.10 We performed immunophenotyping of PB B cells from patients with CLL (n = 118) and age-matched healthy controls (n = 25), to examine the expression of these integrin molecules. Compared with healthy B cells, there was a marked decrease in expression of CD11a, CD11b, CD18, CD29, CD49d, and ITGB7 on CLL cells. However, uniquely among the main cytogenetic categories, CLL cells from patients with trisomy 12 (n = 21) had relatively preserved expression of these integrins, with levels comparable to healthy B cells, although heterogeneity of expression was noted (Figure 1A-F). The expression of a panel of other molecules involved in the leukocyte adhesion cascade was also investigated. The selectins CD162 (PSGL1) and CD62L (l-selectin) are important for the initial capture and rolling of leukocytes, whereas the adhesion molecules CD31 (PECAM-1), CD99, CD321 (JAM-A), and CD323 (JAM-C) mediate paracellular and transcellular leukocyte transmigration. Although the expression of CD31, CD162, and CD321 was increased on CLL cells compared with healthy B cells, there were no differences in the expression of these molecules among the major cytogenetic categories (supplemental Figure 1). The only exception was JAM-C, which while being downregulated on CLL cells in general, was also expressed at a higher level on trisomy 12 cells (P < .01) (supplemental Figure 1F).
Preserved expression of the integrins CD11a, CD11b, CD18, CD29, CD49d, and ITGB7 on trisomy 12 CLL cells. The expression of the integrins CD11a (A), CD11b (B), CD18 (C), CD29 (D), CD49d (E), and ITGB7 (F) were assessed on PB CLL cells and B cells from healthy age-matched controls. Uniquely among the main cytogenetic categories, CLL cells from patients with trisomy 12 had relatively preserved expression of these integrins, with levels comparable to healthy B cells in some patients. A comparable pattern was observed whether the data were analyzed by “% positive” or by median fluorescence intensity.
Preserved expression of the integrins CD11a, CD11b, CD18, CD29, CD49d, and ITGB7 on trisomy 12 CLL cells. The expression of the integrins CD11a (A), CD11b (B), CD18 (C), CD29 (D), CD49d (E), and ITGB7 (F) were assessed on PB CLL cells and B cells from healthy age-matched controls. Uniquely among the main cytogenetic categories, CLL cells from patients with trisomy 12 had relatively preserved expression of these integrins, with levels comparable to healthy B cells in some patients. A comparable pattern was observed whether the data were analyzed by “% positive” or by median fluorescence intensity.
As high expression of the α-integrin CD49d is associated with impaired prognosis in CLL, the prognostic significance of integrin expression was investigated in a cohort of patients from all cytogenetic categories. The characteristics of the patients used for this analysis are summarized in supplemental Table 2. In agreement with previous reports, increased expression of CD49d (>30% positive) was associated with shortened time to first treatment (TTFT) in this cohort (P = .0001).11 Furthermore, increased expression of the other α-integrins CD11a (>11% positive; median expression) and CD11b (>1% positive; threshold set by isotype control) was also associated with a shortened TTFT (CD11a: P = .0025; CD11b: P = .0274) (supplemental Figure 2).
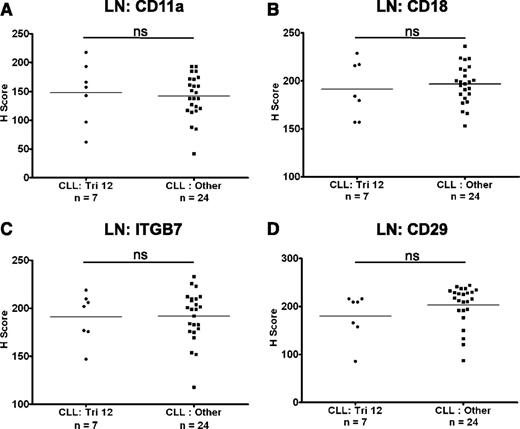
The expression of integrins on CLL cells in LNs
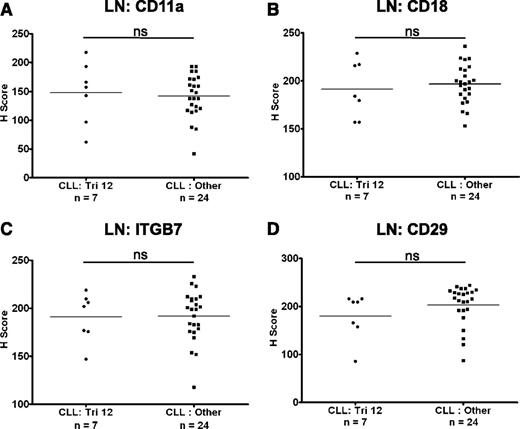
The expression of CD11a, CD18, CD29, and ITGB7 on CLL cells was also investigated in LN biopsies from a cohort of 31 CLL patients with known cytogenetics. The expression of these molecules was heterogeneous on nodal CLL cells from both patients with trisomy 12 (n = 7) and in nontrisomy 12 cases (n = 24). However, in contrast to circulating CLL cells, there was no difference in the expression of CD11a, CD18, CD29, and ITGB7 between these 2 groups (Figure 2A-D). Interestingly, integrin expression on healthy nodal B cells was higher on proliferating germinal center B cells than on mantle zone B cells (Figure 3A), and on proliferating healthy B cells within residual follicles in CLL LNs (supplemental Figure 3). Across diffuse areas of CLL infiltration, increased numbers of proliferating cells were also associated with globally increased CLL-cell expression of CD11a, CD29, and ITGB7 (Figure 3B-C). Although we aimed to characterize the expression of CD49d on nodal B cells, this antigen was not detectable in healthy or CLL LNs with a selection of antibodies, including the clone used for flow cytometric analysis. We conclude that this epitope is destroyed by fixation/paraffin embedding.
The expression of integrins on CLL cells in LNs. The expression of integrins was assessed on nodal CLL cells. In contrast to circulating CLL cells, there was no difference in the expression of CD11a (A), CD18 (B), ITGB7 (C), and CD29 (D) on CLL cells from trisomy 12 and nontrisomy 12 cases.
The expression of integrins on CLL cells in LNs. The expression of integrins was assessed on nodal CLL cells. In contrast to circulating CLL cells, there was no difference in the expression of CD11a (A), CD18 (B), ITGB7 (C), and CD29 (D) on CLL cells from trisomy 12 and nontrisomy 12 cases.
Increased integrin expression correlates with increased numbers of proliferating B cells in healthy and CLL LNs. (A) Representative images of a secondary follicle in a healthy reactive LN. Proliferating germinal center B cells exhibit higher expression of CD11a, CD18, CD29, and ITGB7 than mantle zone B cells. (B) Across LN biopsies from all cytogenetic groups, the presence of higher numbers of proliferating cells correlated with increased expression of CD11a, CD29, and ITGB7, but not CD18. (C) Representative images of CLL LN biopsies without proliferation centers. Biopsies with high numbers of Ki67+ proliferating cells have increased expression of CD11a compared with biopsies with low numbers of Ki67+ proliferating cells. The increased expression of CD11a in biopsies with high numbers of Ki67+ proliferating cells was due to increased staining of the CD79a+ cells.
Increased integrin expression correlates with increased numbers of proliferating B cells in healthy and CLL LNs. (A) Representative images of a secondary follicle in a healthy reactive LN. Proliferating germinal center B cells exhibit higher expression of CD11a, CD18, CD29, and ITGB7 than mantle zone B cells. (B) Across LN biopsies from all cytogenetic groups, the presence of higher numbers of proliferating cells correlated with increased expression of CD11a, CD29, and ITGB7, but not CD18. (C) Representative images of CLL LN biopsies without proliferation centers. Biopsies with high numbers of Ki67+ proliferating cells have increased expression of CD11a compared with biopsies with low numbers of Ki67+ proliferating cells. The increased expression of CD11a in biopsies with high numbers of Ki67+ proliferating cells was due to increased staining of the CD79a+ cells.
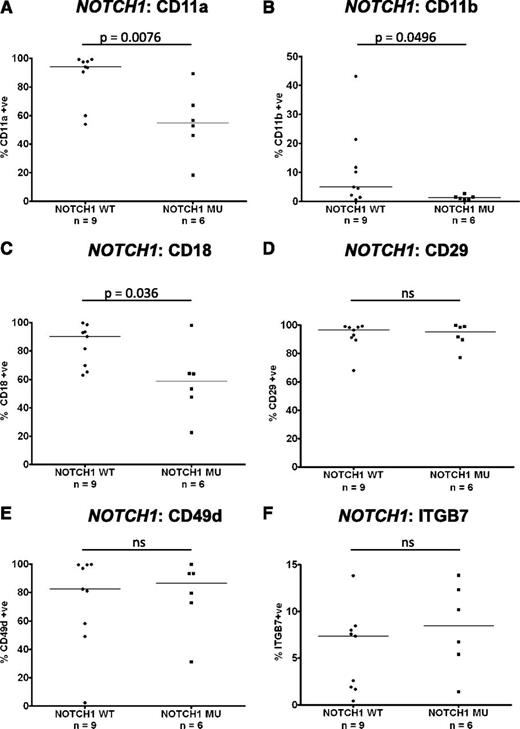
Impact of NOTCH1 mutations on integrin expression in trisomy 12 CLL
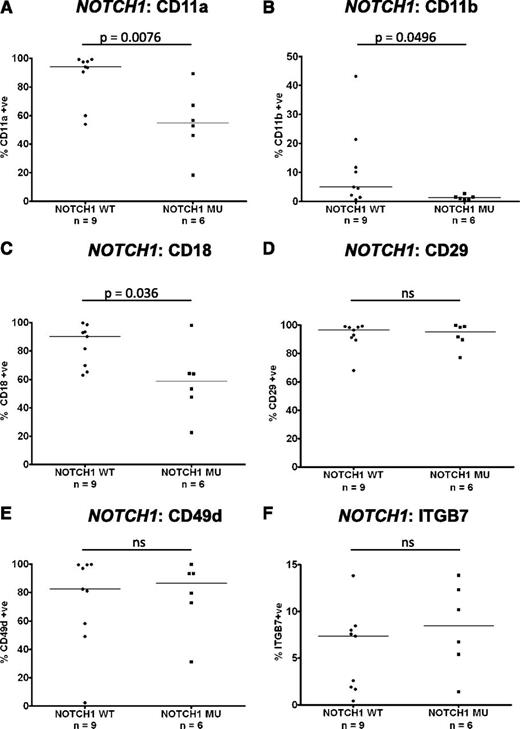
Recent work has identified an association between mutations in the NOTCH1 gene and the presence of trisomy 12.1 The expression of integrins on PB CLL cells with trisomy 12 was compared between cases known to have mutations in NOTCH1 (n = 6) and wild type (n = 9). Interestingly, the presence of a NOTCH1 mutation in the context of trisomy 12 led to decreased CLL-cell expression of CD11a (P = .0076), CD11b (P = .0496), and CD18 (P = .036) to levels comparable with CLL cells without trisomy 12 (Figure 4A-C). Notably, the presence of a NOTCH1 mutation had no impact on CD29, CD49d, or ITGB7 expression (Figure 4D-F). These data suggest that the observed heterogeneity of expression of β2-integrins in trisomy 12 CLL cases is largely explained by the presence of NOTCH1 mutations.
Impact of NOTCH1 mutations on integrin expression in trisomy 12 CLL. The impact of NOTCH1 mutation status on integrin expression was assessed in a cohort of separate cohort of 15 trisomy 12 CLL patients with known NOTCH1 mutation status.1 Notably the expression of CD11a (A), CD11b (B), and CD18 (C) was significantly reduced in trisomy 12 CLL cells with a NOTCH1 mutation compared with trisomy 12 CLL cells with wild-type NOTCH1 genes. NOTCH1 mutation status had no impact on the expression of CD29 (D), CD49d (E), or ITGB7 (F).
Impact of NOTCH1 mutations on integrin expression in trisomy 12 CLL. The impact of NOTCH1 mutation status on integrin expression was assessed in a cohort of separate cohort of 15 trisomy 12 CLL patients with known NOTCH1 mutation status.1 Notably the expression of CD11a (A), CD11b (B), and CD18 (C) was significantly reduced in trisomy 12 CLL cells with a NOTCH1 mutation compared with trisomy 12 CLL cells with wild-type NOTCH1 genes. NOTCH1 mutation status had no impact on the expression of CD29 (D), CD49d (E), or ITGB7 (F).
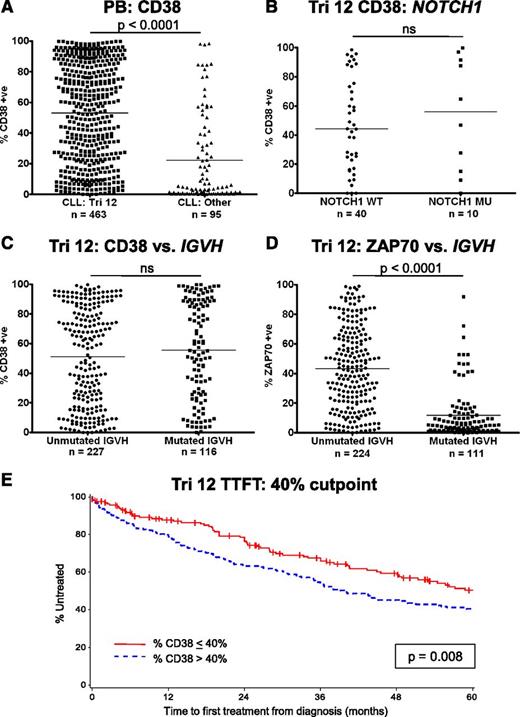
Implications of the increased expression of CD38 on trisomy 12 CLL cells
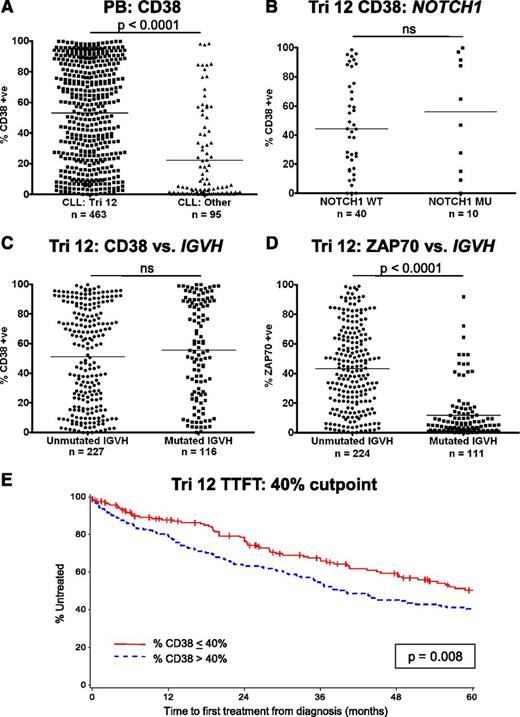
CD38 has several important functions in leukocyte biology, but also acts as an adhesion molecule due to its interactions with CD31 and hyaluronic acid.12,13 High CD38 expression on CLL cells is also a known poor prognostic marker and has been used as a surrogate marker of unmutated IGVH genes.14 In addition, CD38 expression is increased on trisomy 12 CLL cells.5,15 The implications of this observation were investigated in a large cohort of patients with trisomy 12 detectable by fluorescence in-situ hybridization. In agreement with previous reports, CLL cases with trisomy 12 had significantly higher expression of CD38 compared with CLL cells from the other major cytogenetic categories (P < .0001) (Figure 5A). However, mutations in NOTCH1 had no impact on the expression of CD38 (Figure 5B). Strikingly, although there was no correlation of CD38 expression with IGVH mutation status within the trisomy 12 group, the association of unmutated IGVH genes with ZAP70 positivity remained intact (Figure 5C-D).16,17 The impact of the presence of trisomy 12 on prognosis was assessed in a cohort of 422 patients (supplemental Table 3). When the threshold for CD38 positivity was set at the standard 30%, higher expression of CD38 was not associated with a significantly impaired TTFT. Recursive partitioning identified that the optimal cutoff point for TTFT was 42.4%, and using a level of 40% CD38 expression retained its prognostic value for TTFT (P = .008) (Figure 5E). IGVH mutation status and ZAP70 expression retained their prognostic impact in trisomy 12 patients (supplemental Figure 4).
Implications of the increased expression of CD38 on trisomy 12 CLL cells. (A) The proportion of cells that express CD38 is increased in trisomy 12 cases. (B) NOTCH1 mutation status had no impact on the expression of CD38 in trisomy 12 cases. (C) CD38 is not a surrogate marker of IGVH mutation status in patients with trisomy 12. (D) In contrast, increased expression of ZAP70 retains its association with IGVH mutation status in patients with trisomy 12. Treatment-free survival curves for CLL patients with trisomy 12 with a 40% cutoff for CD38 positivity (E).
Implications of the increased expression of CD38 on trisomy 12 CLL cells. (A) The proportion of cells that express CD38 is increased in trisomy 12 cases. (B) NOTCH1 mutation status had no impact on the expression of CD38 in trisomy 12 cases. (C) CD38 is not a surrogate marker of IGVH mutation status in patients with trisomy 12. (D) In contrast, increased expression of ZAP70 retains its association with IGVH mutation status in patients with trisomy 12. Treatment-free survival curves for CLL patients with trisomy 12 with a 40% cutoff for CD38 positivity (E).
Integrin “inside-out” signaling is upregulated in trisomy 12 CLL cells
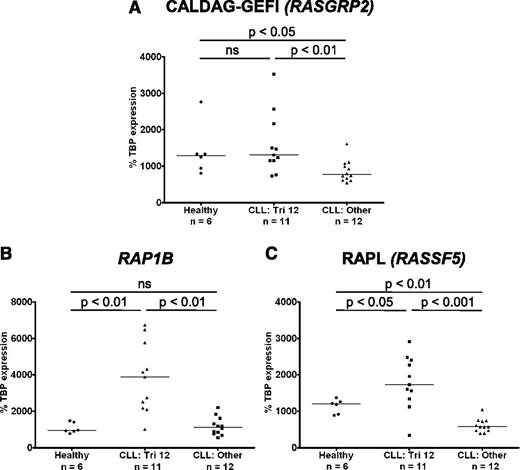
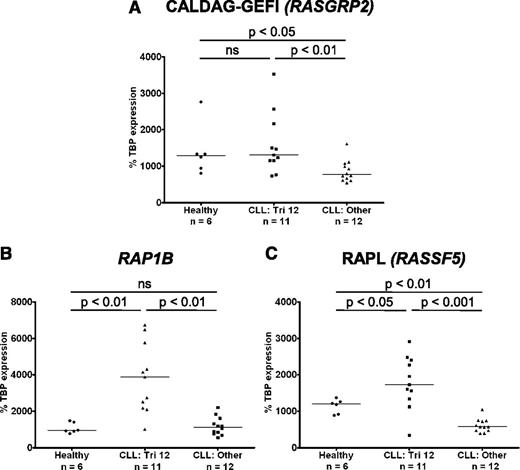
In light of the upregulation of integrins on trisomy 12 CLL cells, the expression of molecules involved in integrin “inside-out” signaling was investigated. Binding of ligand to G-protein coupled receptors results in activation of intracellular signaling cascades and increases in cytosolic calcium and diacylglycerol (DAG). These are sensed by the guanine-nucleotide exchange factor (GEF) calcium- and DAG- regulated GEFI (CALDAG-GEFI; RASGRP2), which in turn activates the small GTPase Ras-related protein (RAP1).10 Notably, the gene RAP1B, the dominant isoform of RAP1 in B lymphocytes, is coded for on chromosome 12. RAP1 can then either activate integrins directly, or through its intermediate effector Ras-related protein ligand (RAPL) (RASSF5).
The expression of CALDAG-GEFI, RAP1B, and RAPL was investigated by RT-PCR. Importantly CALDAG-GEFI expression was significantly higher in CLL cells with trisomy 12 than in nontrisomy 12 cases, and levels of expression were comparable to those in healthy B cells (Figure 6A). Furthermore, both RAP1B and its effector RAPL were overexpressed in trisomy 12 CLL cells compared with both healthy B cells, and CLL cells without trisomy 12 (Figure 6B-C). In contrast, there was no difference in RAP1A expression when comparing healthy B cells with CLL cells or between the different cytogenetic groups. Interestingly, although the expression of the signal transduction adaptor paxillin was upregulated in CLL cells and the structural molecules talin and vinculin were downregulated, there was no difference among the cytogenetic groups (supplemental Figure 5).
Integrin “inside-out” signaling is upregulated in trisomy 12 CLL cells. The expression of molecules involved in integrin signaling was assessed by quantitative RT-PCR in CLL cells with and without trisomy 12 and healthy B cells. (A) The expression of RASGRP2 (CALDAG-GEFI) is increased in trisomy 12 CLL cells comparable to healthy B cells. In contrast, RAP1B (B) and RASSF5 (RAPL) (C) are overexpressed in trisomy 12 CLL cells compared with healthy B cells and nontrisomy 12 CLL cells.
Integrin “inside-out” signaling is upregulated in trisomy 12 CLL cells. The expression of molecules involved in integrin signaling was assessed by quantitative RT-PCR in CLL cells with and without trisomy 12 and healthy B cells. (A) The expression of RASGRP2 (CALDAG-GEFI) is increased in trisomy 12 CLL cells comparable to healthy B cells. In contrast, RAP1B (B) and RASSF5 (RAPL) (C) are overexpressed in trisomy 12 CLL cells compared with healthy B cells and nontrisomy 12 CLL cells.
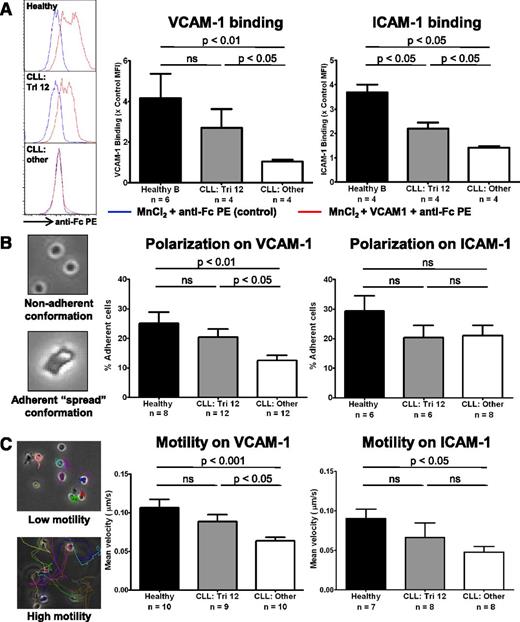
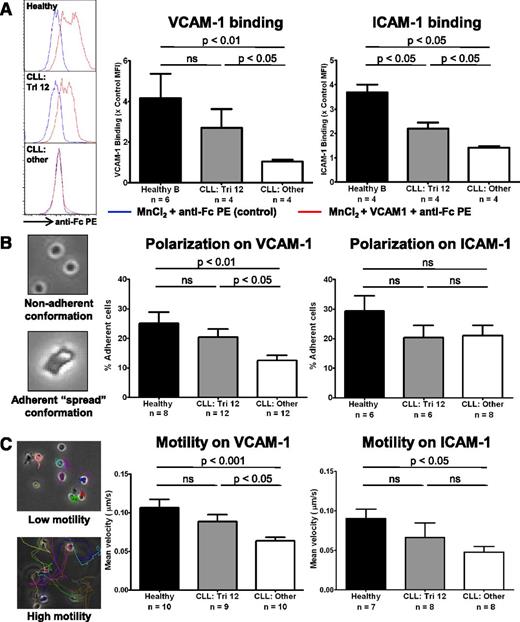
The upregulation of integrin expression results in increased ligand binding, and enhanced adhesion and motility that is predominantly VLA-4 directed
The functional consequence of upregulated integrin expression on trisomy 12 CLL cells was evaluated. MnCl2 was used to induce integrin conformational changes to establish whether increased expression of VLA-4 and LFA-1 integrins resulted in enhanced ability to bind their respective ligands VCAM-1 and ICAM-1.18 Although healthy B cells were able to bind significant amounts of ligand, nontrisomy 12 CLL cells bound very little VCAM-1 or ICAM-1 after MnCl2 treatment, with trisomy 12 CLL cells intermediate between the 2 (Figure 7A). Next, we tested whether the increased integrin expression resulted in an enhanced ability to adhere to and polarize on immobilized VCAM-1 and ICAM-1 after stimulation by CXCL12 (SDF1α). This also examined the ability of the cells to adopt a spread adherent conformation, reflecting cytoskeletal function. After stimulation with CXCL12, trisomy 12 CLL cells exhibited enhanced adherence to plate-bound VCAM-1 comparable to healthy B cells. However, there was no improvement in adherence to ICAM-1 (Figure 7B and supplemental Figure 6). Then we performed migration tracking of CLL cells from patients with or without trisomy 12 compared with healthy B cells to assess the effects of the differences in integrin expression on cell motility after stimulation with CXCL12. Mirroring the adhesion data, trisomy 12 CLL cells exhibited increased motility on plate-bound VCAM-1 compared with the other cytogenetic groups, with an average velocity comparable to healthy B cells. However, there was no significant difference in motility on ICAM-1 in the trisomy 12 group (Figure 7C and supplemental Figure 6). Together, this data indicates that the increased expression of β1-integrins on trisomy 12 CLL is functionally significant and results in enhanced adhesion and motility that is predominantly VLA-4 mediated.
The upregulation of integrin signaling results in increased ligand binding and enhanced adhesion and motility that is predominantly VLA-4 directed. The integrin function of CLL cells and healthy B cells was investigated. (A) The ability of the cells to bind soluble VCAM-1 or ICAM-1 was assessed by flow cytometry after integrin activation by 3 mM MnCl2. (A) Healthy B cells (n = 4) are able to bind significant amounts after VCAM-1 and ICAM-1 after integrin activation, whereas nontrisomy 12 CLL cells (n = 4) bind comparatively little. Trisomy 12 CLL cells (n = 4) bind an intermediate amount of these ligands consistent with their increased integrin expression. The adhesive ability and nondirectional motility of healthy and malignant B cells on VCAM-1– and ICAM-1–coated plates was examined. (B) The proportion of cells in a “spread” conformation was assessed 30 minutes after stimulation with CXCL12. Trisomy 12 CLL cells exhibit an enhanced ability to adhere to immobilized VCAM-1, but not immobilized ICAM-1. (C) This enhanced adhesion translates into improved motility on VCAM-1, but was not significantly increased on ICAM-1. Error bars in all figures represent standard error of the mean.
The upregulation of integrin signaling results in increased ligand binding and enhanced adhesion and motility that is predominantly VLA-4 directed. The integrin function of CLL cells and healthy B cells was investigated. (A) The ability of the cells to bind soluble VCAM-1 or ICAM-1 was assessed by flow cytometry after integrin activation by 3 mM MnCl2. (A) Healthy B cells (n = 4) are able to bind significant amounts after VCAM-1 and ICAM-1 after integrin activation, whereas nontrisomy 12 CLL cells (n = 4) bind comparatively little. Trisomy 12 CLL cells (n = 4) bind an intermediate amount of these ligands consistent with their increased integrin expression. The adhesive ability and nondirectional motility of healthy and malignant B cells on VCAM-1– and ICAM-1–coated plates was examined. (B) The proportion of cells in a “spread” conformation was assessed 30 minutes after stimulation with CXCL12. Trisomy 12 CLL cells exhibit an enhanced ability to adhere to immobilized VCAM-1, but not immobilized ICAM-1. (C) This enhanced adhesion translates into improved motility on VCAM-1, but was not significantly increased on ICAM-1. Error bars in all figures represent standard error of the mean.
Discussion
Transendothelial migration of leukocytes is a complex process mediated by the concerted activity of selectins, integrins, adhesion molecules, and chemokines.10 Here, we investigated expression of a range of molecules implicated in the leukocyte adhesion cascade. Although we observed that the expression of the integrins CD11a, CD11b, CD18, CD29, CD49d, and ITGB7 was decreased on circulating CLL cells in general, uniquely among the main cytogenetic categories, their expression was relatively preserved on trisomy 12 CLL cells. These differences in surface integrin expression were associated with upregulation of molecules involved in intracellular integrin signaling. These changes were of functional significance, as trisomy 12 CLL cells exhibited increased ICAM-1 and VCAM-1 binding on integrin activation, and showed enhanced VLA-4-mediated adhesion and motility. The effect of trisomy 12 was dominant, with upregulation of integrin signaling also present in trisomy 12 with other cytogenetic abnormalities including del-11q or del-17p. Whereas increased CD38 expression with trisomy 12 has been previously reported,5,15 its prognostic significance has not been evaluated. Importantly, we demonstrate that CD38 cannot be used as a surrogate marker of IGVH gene mutation status in this subgroup and the threshold of CD38 positivity should be raised to 40% in the presence of trisomy 12 for this marker to retain its prognostic value.
An enhanced ability for trisomy 12 CLL cells to undergo transendothelial migration may account for some of the clinical characteristics associated with the presence of this cytogenetic abnormality. The increased prevalence of trisomy 12 in SLL and Richter’s transformation may reflect enhanced ability of CLL cells to migrate into LNs, resulting in a shift in disease distribution from the leukemic phase into the LNs. Notably, there was no difference in expression of integrins between trisomy 12 and nontrisomy 12 CLL cells within LNs. This could be the result of several different factors. First, relatively high integrin expression could be required for CLL cells to enter LNs, and hence nodal CLL cells are selected for their higher expression of these molecules. Second, CLL cells are known to encounter several different survival and proliferation signals with the LN microenvironment, which may lead to upregulation of integrin expression. Finally, this may also represent a bias in sampling because although the PB samples were taken at all stages prior to initial therapy, the LNs were usually biopsied immediately prior to treatment at a more advanced disease stage.
We hypothesized that increased expression of molecules involved in the leukocyte adhesion cascade would result in increased homing to tissue microenvironments, whereas CLL cells would encounter more survival and proliferation signals, potentially resulting in more aggressive disease. This hypothesis appeared to be borne out across the cytogenetic categories, with increased expression of CD11a and CD11b associated with shortened TTFT, in addition to increased CD49d. Increased CD11a, CD29, and ITGB7 expression also correlated with higher numbers of proliferating CLL cells in LNs, reflecting normal B-cell biology. A paradoxical finding from this study is that despite the trisomy 12 group having the highest expression of integrins and enhanced function, this cytogenetic abnormality confers intermediate prognosis.19 Despite having a large cohort of trisomy 12 patients, none of the analyses regarding overall survival and CD38 expression reached statistical significance due to the relatively few deaths observed in this group. Therefore, although increased interaction with the tissue microenvironment does confer a negative prognosis, other factors, such as the genomic instability associated with loss of 17p or 11q are clearly more important. A particularly interesting observation was the interplay between NOTCH1 mutational status and integrin expression. Although the presence of a NOTCH1 mutation with trisomy 12 led to decreased expression of the β2-integrins CD11a/CD18 (LFA-1) and CD11b/CD18 (Mac-1) on CLL cells, NOTCH1 mutations had no impact on CD29, CD49d, ITGB7, or CD38 expression. This division is of functional importance as the α-integrin CD49d pairs with either of the β-integrins (CD29 or ITGB7) to form integrin dimers, and this forms a macromolecular cell surface complex with CD38, CD44, and matrix metalloproteinase 9 on CLL cells.20,21 Importantly, these findings suggest that our results are not consistent with increased motility contributing to the adverse prognosis associated with NOTCH1 mutations, as differential β2-integrin expression was not associated with any LFA-1–mediated functional changes in our assays. It is possible that other functional effects may be important, and we hypothesize that NOTCH1-induced suppression of β2-integrin expression may allow escape from immune surveillance. At this time, there is little published data regarding direct interactions between NOTCH signaling and integrin expression. However, it is possible that the increased NOTCH activity in NOTCH1 mutated cases leads to inhibition of β2-integrin-transcription via a NOTCH target gene.22 For example, overexpression of c-Myc has previously associated with reduced β2-integrin expression, with this transcription factor being shown to have a direct effect on integrin gene transcription.23 This interaction between NOTCH1 and β2-integrin signaling pathways is an important area of future investigation in attempts to understand the role of NOTCH1 mutations in aggressive CLL.
Our findings also have implications for our understanding of CLL cell motility and behavior. Although increased expression of CD29/CD49d (VLA-4) resulted in enhanced adhesion and motility on VCAM-1 coated plates, increased expression of CD11a/CD18 (LFA-1) did not result in significantly enhanced adhesion and motility on ICAM-1, despite improved ligand binding. Interestingly, the transmigratory capacity of CLL cells varies among patients, with CLL cells from patients with advanced disease and lymphadenopathy having increased rates of transendothelial migration. Importantly, increased expression of CCR7 and VLA-4 are key factors in this enhanced migration, with levels of CD49d expression correlating with the presence of lymphadenopathy.24 A similar association has also been shown between high expression of CD49d and increased bone marrow infiltration in human disease, and enhanced bone marrow homing capacity in an in vitro adoptive transfer mouse model.25 Mechanistically, there is evidence to suggest that while entry of normal B cells into LNs is dependent on LFA-1, CLL cells rely on interactions between VLA-4 and LFA-1 to cross endothelial cell monolayers.26,27 Taken together, the evidence suggests that VLA-4 plays a more important role than LFA-1 in the migratory function of CLL cells, which is also being borne out in novel models of CLL cell trafficking.28,29
In addition to the importance of integrin expression on CLL cell migration, changes in intracellular signaling have also been demonstrated to play a role in CLL cell migration. Impaired chemokine responsiveness in CLL cells is likely due to a failure of Rap1-containing endosomes to translocate to the plasma membrane, resulting in defective in Rap1 GTP-loading.30,31 It is possible that overexpression of CALDAG-GEFI, RAP1B, and RAPL in trisomy 12 CLL cells may compensate for this defect and further improve function, and this will be the focus of future experiments. The mechanisms underlying upregulation of integrin signaling in trisomy 12 remain unclear, although a recent report has implicated altered epigenetic regulation as a cause of increased CD49d expression.6 The presence of an extra copy of chromosome 12 may affect gene expression, and it is notable that the genes encoding both RAP1B and ITGB7 are located on chromosome 12. However, the genes for the other integrins and signaling molecules are located elsewhere in the genome, and molecules such as paxillin were not significantly upregulated in trisomy 12 despite also being encoded on chromosome 12.
In conclusion, we demonstrate that trisomy 12 CLL cells exhibit enhanced expression of integrin signaling molecules compared with the other cytogenetic groups. These changes are associated with enhanced function that may account for the unique clinical characteristics of this group. Importantly the expression of the β2-integrins CD11a/CD18 (LFA-1) and CD11b/CD18 (Mac-1) are downregulated by the coexistence of NOTCH1 mutations, indicating a novel interaction that may be of potential importance in aggressive poor risk CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and healthy controls who donated their blood and tissue.
This work was supported by grants from Cancer Research UK (J.C.R. and J.G.G.), the European Hematology Association (A.G.R.), and by funding from the National Cancer Institute (P01 CA95426; J.G.G., C.M.C., L.Z.R., L.W., D.S.N., and T.J.K.) for the CLL Research Consortium and from Goldman Sachs (J.C.R. and J.G.G.).
Authorship
Contribution: J.C.R. designed and performed the experiments, analyzed and interpreted the data, and wrote the manuscript; A.J.C., C.J.D., S.J.K., F.M., and A.G.R. designed and performed the experiments and analyzed the data; C.M.C., L.Z.R., and T.J.K. provided the samples and the CD38 and NOTCH1 data, and edited the manuscript; D.S.N. and L.W. analyzed and interpreted the data, and edited the manuscript; and J.G.G. designed the experiments, interpreted the data, wrote and edited the manuscript, and supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John C. Riches, Barts Cancer Institute, Queen Mary University of London, 3rd Floor John Vane Science Centre, Charterhouse Square, London, EC1M 6BQ United Kingdom; e-mail: johnriches@doctors.org.uk.