Key Points
Neutrophil lifespan is extended in patients with gain-of-function HIF2A mutations.
HIF-2α regulates in vivo neutrophil longevity and thus tissue inflammation and repair.
Abstract
Neutrophil lifespan and function are regulated by hypoxia via components of the hypoxia inducible factor (HIF)/von Hippel Lindau/hydroxylase pathway, including specific roles for HIF-1α and prolyl hydroxylase-3. HIF-2α has both distinct and overlapping biological roles with HIF-1α and has not previously been studied in the context of neutrophil biology. We investigated the role of HIF-2α in regulating key neutrophil functions. Human and murine peripheral blood neutrophils expressed HIF-2α, with expression up-regulated by acute and chronic inflammatory stimuli and in disease-associated inflammatory neutrophil. HIF2A gain-of-function mutations resulted in a reduction in neutrophil apoptosis both ex vivo, through the study of patient cells, and in vivo in a zebrafish tail injury model. In contrast, HIF-2α–deficient murine inflammatory neutrophils displayed increased sensitivity to nitrosative stress induced apoptosis ex vivo and increased neutrophil apoptosis in vivo, resulting in a reduction in neutrophilic inflammation and reduced tissue injury. Expression of HIF-2α was temporally dissociated from HIF-1α in vivo and predominated in the resolution phase of inflammation. These data support a critical and selective role for HIF-2α in persistence of neutrophilic inflammation and provide a platform to dissect the therapeutic utility of targeting HIF-2α in chronic inflammatory diseases.
Introduction
Neutrophils are key mediators of tissue injury in acute and chronic inflammatory diseases.1,2 Timely neutrophil apoptosis, with effective macrophage efferocytosis, ensures resolution of inflammation and protects against the cytotoxic effects of neutrophils.3-5 As such, targeting neutrophil apoptosis represents an attractive therapeutic strategy.
Neutrophil apoptosis is significantly inhibited by physiological hypoxia.6-8 Adaptation to hypoxia is mediated by the heterodimeric nuclear transcription factor, hypoxia inducible factor (HIF), comprised of 1 of 3 known α subunits and a constitutively expressed β subunit.9 In normoxia, HIF is inactive as α subunits are labeled for degradation by a family of 3 oxygen-sensitive prolyl hydroxylases (PHDs), and HIF transcription may be inactivated by factor inhibiting HIF.10-12
Recent evidence reveals the importance of oxygen-sensing pathways in innate immune biology. HIF-α subunits accumulate in myeloid cells in hypoxic conditions, as in other cell types, but also in response to bacteria and bacterial products irrespective of the ambient oxygen tension. These data demonstrate roles for the HIF pathway beyond the regulation of hypoxic signaling and implicate HIF in host responses to bacteria.13-15 Indeed, myeloid-specific deficiency of HIF-1α not only abolishes the prolonged survival of neutrophils in hypoxia but also results in depletion of intracellular ATP levels and impairment of neutrophil granule protease production, macrophage motility and invasion, and bacterial killing.6,13,16 These in vitro findings translated into reduced inflammatory cell infiltrates in murine models of inflammation and infection.13,16 With such profound effects on innate immune cell function, HIF-1α itself is not an attractive therapeutic target for the many inflammatory diseases, eg, chronic obstructive pulmonary disease (COPD) and inflammatory bowel disease, where inflammation and bacteria frequently coexist. In marked contrast to HIF-1α deficiency, we found that deficiency of a HIF hydroxylase, PHD3, had minimal consequences for the functional status of neutrophils prior to their apoptosis.17 Specific targeting of individual components of the HIF hydroxylase pathway, independent of HIF-1α itself, could therefore result in selective regulation of neutrophil survival pathways independent of key host-pathogen responses.
Distinct biological roles for HIF-1α and HIF-2α have recently emerged, with HIF-2α regulating a distinct but overlapping set of target genes to HIF-1α and, importantly, playing a less significant role in regulating glycolytic enzyme expression.18,19 Furthermore, differential transcriptional activation of HIF-1α and HIF-2α can result in coordinated cellular responses, dependent on the relative abundance of each isoform, with HIF-1α and HIF-2α having opposing effects on macrophage nitric oxide formation.20 HIF-2α has been implicated in regulation of other macrophage functions, with myeloid-specific HIF-2α–deficient mice having reduced macrophage-mediated inflammatory responses to endotoxemia and reduced tumor-associated macrophage infiltration associated with reduced tumor cell proliferation and progression.14 Given this evidence of differential functions for HIF-1α and HIF-2α in myeloid cells and the known dominance of HIF-1α over HIF-2α in the regulation of glycolysis and ATP generation, we hypothesized that HIF-2α deficiency may have a more selective immunomodulatory phenotype than HIF-1α deficiency in neutrophils.
Materials and methods
Ethical approval
All participants gave written informed consent in accordance with the Declaration of Helsinki principles. South Sheffield Research Ethics Committee approved the study of healthy volunteers and individuals with inflammatory arthritis, Oxfordshire Clinical Research Ethics Committee approved the study of patients with gain-of-function mutations in HIF2A, and Helsinki University Hospital, Helsinki, Finland, approved the use of human lung tissue.
Murine colonies
Lysozyme M-driven cre recombinase (LysMcre) was used to target Hif1a (Hif1aflox/flox;LysMcre+/−) or Hif2a (Hif2aflox/flox;LysMcre+/−) deletions in myeloid lineage cells. Animals were back-crossed to a C57BL/6 background.16,20 C57BL/6 mice or littermate LysMcre−/− floxed mice were used as controls. All animal experiments were conducted in accordance with the Home Office Animals (Scientific Procedures) Act of 1986 with local ethics approval.
Isolation and culture of neutrophils from humans and mice
Human peripheral blood neutrophils were isolated from whole blood using dextran sedimentation and discontinuous plasma-Percoll gradients.21 Ultrapurified human neutrophils were obtained using negative magnetic selection as previously described.22 Murine peripheral blood neutrophils and bone marrow–derived neutrophils were isolated by negative magnetic selection (Easysep; STEMCELL Technologies, Grenoble, France), and inflammatory neutrophils were recovered from bronchoalveolar lavage (BAL) fluid 24 hours following challenge with nebulized lipopolysaccharide (LPS). Cell culture is detailed in the supplemental Methods on the Blood Web site.
RNA isolation and relative quantification
Human neutrophils (10 × 106/condition) were lysed with 1 mL TRI Reagent (Sigma-Aldrich Ltd., Gillingham, United Kingdom), and RNA was extracted using chloroform phase partitioning and isopropanol. Murine peripheral blood, inflammatory (BAL), or bone marrow neutrophils (1 × 106/condition) were lysed, and RNA was extracted using the mirVana total RNA isolation protocol (Ambion, Austin, TX). Samples were treated with DNase (Ambion) and random hexamer cDNA synthesized by reverse transcription. Assays-on-demand gene expression TaqMan MGB 6FAM dye-labeled products (Applied Biosystems) were used for relative quantification of cDNA and BigDye v3.1 sequencing kits for product verification (supplemental Methods).
Immunoblot detection of human and murine neutrophil protein
Whole cell human (hypotonic) and murine (sodium dodecyl sulfate [SDS]) lysates were prepared as previously described.17 Immunoblotting was performed with polyclonal anti-mouse HIF-1α (R&D), monoclonal anti-human HIF-1α (clone 54, BD,) or anti–HIF-2α (clone ep190b; Novus Biologicals) primary antibodies. Sample loading was confirmed by p38 mitogen-activated protein kinase (MAPK) expression (Cell Signaling Technology). All bands shown were at the predicted molecular weight for the protein of interest.
Immunohistochemistry
Lung tissue sections were from nonsmokers and patients with mild (The Global Initiative for Chronic Obstructive Lung Disease stage 2) and moderate (The Global Initiative for Chronic Obstructive Lung Disease stage 3/4) COPD undergoing resection for suspected lung tumor or lung transplantation from the Department of Medicine and Pathology, Helsinki University Hospital. Slides were stained with anti–HIF-2α (clone ep190b; Novus Biologicals) or isotype control, developed using the ImmPRESS universal polymer detection kit (Vector Laboratories Ltd, Peterborough, United Kingdom) and visualized with diaminobenzidine.
Neutrophil functional assays
Phagocytosis.
Uptake of Alexa Fluor 488 Escherichia coli (K-12 strain) BioParticles (Invitrogen, Paisley, United Kingdom) after 30 minutes of coculture (multiplicity of infection of 1:1) was determined by flow cytometry (FACSCalibur; Becton Dickinson). Alternatively, neutrophils were incubated with opsonized zymosan (0.2-1 mg/mL) for 30 minutes, and the phagocytic index was calculated using microscopy.
Respiratory burst.
Cells were cultured with 6 µM 2′,7′-dichlorofluorescin diacetate (Sigma-Aldrich) for 30 minutes, and then stimulated for a further 30 minutes with N-formyl-Met-Leu-Phe (100 nM) or opsonized zymosan (0.2 mg/mL) before FL1 fluorescence was determined by flow cytometry and geometric mean fluorescence calculated using FlowJo software (Tree Star Inc.).
Fish husbandry
The neutrophil specific fluorescent zebrafish line Tg(mpx:GFP)i114 was used, subsequently referred to as mpx:GFP for simplicity.23 Zebrafish were maintained according to standard protocols.24 Adult fish were maintained on a 14-hour light and 10-hour dark cycle at 28°C in United Kingdom Home Office–approved facilities in the Medical Research Council Centre for Developmental and Biomedical Genetics aquaria at the University of Sheffield.
Fish assays
Inflammatory responses were elicited in zebrafish larvae by tail transection as previously described, using the neutrophil specific line, mpx:GFP.23,25 Rates of apoptosis were assessed using terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling/tyramide signal amplification, by blinded assessors, and by anti-active caspase-3/tyramide signal amplification staining, as previously described.25 Collagen formation following tailfin injury was assessed at 24, 48, or 72 hours postinjury (hpi). Wild-type zebrafish were fixed and stained with an anti-collagen1 primary antibody and Alexa Fluor 488–conjugated secondary antibody. Fluorescence intensity was analyzed by Volocity 5 (Improvision; Perkin Elmer).
Wild-type and mutant hif2a cloning
Zebrafish 2dpf RNA purified using TRIzol (Invitrogen) was used for reverse transcriptase-polymerase chain reaction (PCR) cloning of HIF2A homologs, hif2aa (epas1a) and hif2ab (epas1b) (primer details in supplemental Table 1), using Pfusion polymerase (Finnzymes, Espoo, Finland). These were initially cloned into the TOPOBlunt vector (Invitrogen) and subsequently subcloned into the pCS2+ vector (Invitrogen) for RNA synthesis. Dominant active forms of hif2aa and hif2ab were generated by successive rounds of site-directed mutagenesis as previously described.25 The zebrafish amino acid corresponding to human HIF-2α G537R and G537W was mutated in the same fashion. RNA encoding dominant active isoforms were transcribed (mMessageMachine; Ambion, Life Technologies) and microinjected into zebrafish embryos at the 1-cell stage.25 To assess the function of overexpressed G487 mutant constructs, phd3 in situ hybridization was performed 24 hours following injection of RNA for G487R, G487W or dominant active hif2aa at the 1-cell stage.26 A dominant-negative form of hif2aa was generated using primers amplifying DNA corresponding to amino acids 1-330 of human HIF-2α.25,27
Murine LPS acute lung injury model
Nebulized LPS (3 mg) was administered to awake mice. At specified time points, BAL sampling was performed and then analyzed for total (hemocytometer), differential, and apoptosis (cytospin) cell counts. BAL supernatant and plasma were stored for cytokine analysis. For histological sections, lungs were fixed with 10% formalin at 20 cm H2O and paraffin-embedded blocks were prepared. Following deparaffinization, serial sections were stained with anti–HIF-2α (clone ep190b).
Bone marrow transplantation
C57BL/6 recipient mice were irradiated with 3 fractions of 1 Gy each day for 4 days before injection with 1.5 × 106 bone marrow cells from wild-type Hif2aflox/flox;LysMcre−/− or knockout Hif2aflox/flox;LysMcre+/− mice.28 Acute lung injury experiments were performed 5 weeks following injection of donor marrow, and reconstitution was confirmed by genotyping of cells recovered from BAL (DNeasy; Qiagen, Crawley, United Kingdom).
Statistical analysis
Data were analyzed using Prism 5.0 software (GraphPad Software, San Diego, CA) using unpaired, 2-tailed t-tests for comparisons between 2 groups and 1-way or 2-way analysis of variance (ANOVA; with Bonferonni post-test adjustment) for other data as appropriate. Significance was accepted when P < .05. Data are expressed as mean ± standard error of the mean (SEM).
Results
Human neutrophils express HIF-2α
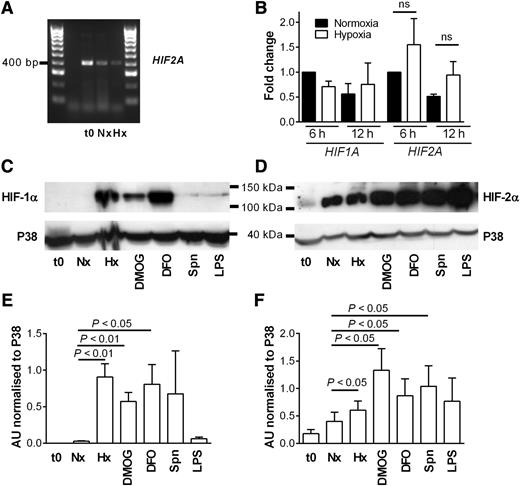
We isolated human peripheral blood neutrophils and demonstrated HIF2A mRNA expression by nonquantitative PCR in highly purified neutrophil populations (Figure 1A), with sequencing of PCR products (data not shown). Real-time PCR assays showed HIF2A expression is not altered over time in normoxic or hypoxic culture (Figure 1B) or following stimulation by heat-killed Staphylococcus aureus or peptidoglycan (supplemental Figure 1). In contrast to HIF-1α, HIF-2α protein was detected in freshly isolated neutrophils and neutrophils cultured in normoxia, with further induction of HIF-2α protein and expression of HIF-1α protein in hypoxia. HIF-2α was up-regulated with iron chelators, hydroxylase inhibitors, heat-killed bacteria, or LPS (Figure 1C-F).
Human neutrophils express HIF-2α, and expression is up-regulated by hypoxia, hydroxylase inhibition, and heat-killed bacteria. (A) Expression of HIF2A in freshly isolated neutrophils (t0) or cells cultured for 4 hours in normoxia (N) or hypoxia (H) was determined by PCR and agarose gel electrophoresis. A representative gel image of n = 3 is shown. (B) Fold change in expression of HIF2A and HIF1A following culture of human neutrophils in normoxia (filled bars) or hypoxia (open bars) for 6 or 12 hours. TaqMan analysis of cDNA was performed with data normalized to ACTB expression. Data show mean and SEM of fold change with respect to normoxic samples at 6 hours, n = 3, analyzed by ANOVA. (C-D) Expression of HIF-1α and HIF-2α is differentially regulated by hydroxylase inhibitors and up-regulated in response to heat-killed bacteria. Neutrophils were cultured in normoxia (Nx) with dimethyloxalylglycine (DMOG; 100 µM); deferoxamine (DFO; 300 µM); heat-killed Streptococcus pneumoniae (Spn; multiplicity of infection 10:1); LPS (100 ng/mL); or hypoxia (Hx) before being lysed. Proteins were separated using SDS-polyacrylamide gel electrophoresis (PAGE), and blots were probed for (C) HIF-1α and (D) HIF-2α. p38 MAPK was used as a loading control. (E-F) Densitometry analysis was performed on (E) HIF-1α and (F) HIF-2α blots using ImageJ software and normalized to p38 MAPK expression. Data are mean and SEM for minute n = 4.
Human neutrophils express HIF-2α, and expression is up-regulated by hypoxia, hydroxylase inhibition, and heat-killed bacteria. (A) Expression of HIF2A in freshly isolated neutrophils (t0) or cells cultured for 4 hours in normoxia (N) or hypoxia (H) was determined by PCR and agarose gel electrophoresis. A representative gel image of n = 3 is shown. (B) Fold change in expression of HIF2A and HIF1A following culture of human neutrophils in normoxia (filled bars) or hypoxia (open bars) for 6 or 12 hours. TaqMan analysis of cDNA was performed with data normalized to ACTB expression. Data show mean and SEM of fold change with respect to normoxic samples at 6 hours, n = 3, analyzed by ANOVA. (C-D) Expression of HIF-1α and HIF-2α is differentially regulated by hydroxylase inhibitors and up-regulated in response to heat-killed bacteria. Neutrophils were cultured in normoxia (Nx) with dimethyloxalylglycine (DMOG; 100 µM); deferoxamine (DFO; 300 µM); heat-killed Streptococcus pneumoniae (Spn; multiplicity of infection 10:1); LPS (100 ng/mL); or hypoxia (Hx) before being lysed. Proteins were separated using SDS-polyacrylamide gel electrophoresis (PAGE), and blots were probed for (C) HIF-1α and (D) HIF-2α. p38 MAPK was used as a loading control. (E-F) Densitometry analysis was performed on (E) HIF-1α and (F) HIF-2α blots using ImageJ software and normalized to p38 MAPK expression. Data are mean and SEM for minute n = 4.
Overexpression of HIF-2α delays neutrophil apoptosis but does not affect function
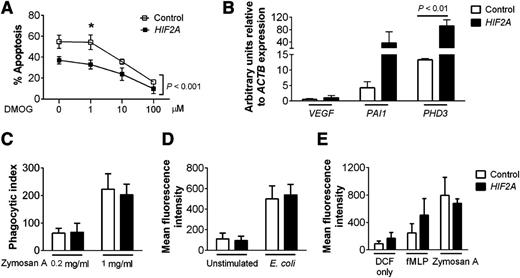
Study of idiopathic cases of erythrocytosis with raised serum erythropoietin has resulted in the identification of a rare group of individuals with gain-of-function mutations in the HIF2A gene.29 We obtained peripheral blood neutrophils from some of these individuals to determine the consequences of HIF-2α overexpression for neutrophil survival and function. Subjects with gain-of-function HIF2A mutations had lower rates of neutrophil apoptosis compared with controls but a preserved response to hydroxylase inhibition by the pan-hydroxylase inhibitor dimethyloxalylglycine (DMOG) (Figure 2A). Neutrophils from these subjects also showed enhanced expression of the HIF2A target genes PAI1 and PHD3 (Figure 2B), which were equivalent to changes previously described following hypoxic culture.17 Functional assays showed phagocytosis (Figure 2C-D) and respiratory burst (Figure 2E) were equivalent between control and HIF2A mutant neutrophils, indicating HIF-2α overexpression resulted in a selective prosurvival phenotype without alteration of key neutrophil functions.
Neutrophils isolated from patients with gain-of-function HIF2A mutations have enhanced survival but normal function. (A) Apoptosis. Neutrophils from patients with HIF2A mutations (closed squares) or healthy controls (open squares) were cultured for 20 hours with dimethyloxalylglycine (0-100 µM) and apoptosis was determined by morphology. Data are mean ± SEM for n = 3, analyzed by 2-way ANOVA with Bonferroni’s multiple comparison post-test. Overall, there was a significant difference between the control group and the patient group (P < .001), with multiple comparison testing showing statistical significance at the 1 µM concentration (*P < .05). (B) Expression of the HIF targets VEGF, PAI1, and PHD3. TaqMan quantitative PCR analysis of cDNA prepared from freshly isolated neutrophils of patients with HIF2A mutations (filled bars) or healthy controls (open bars). Data are mean and SEM for n = 3. (C-D) Phagocytosis. (C) Phagocytic index was calculated from cytospin slides of neutrophils from healthy controls (open bars) and HIF2A patients (filled bars) prepared after 30 minutes of culture with opsonized zymosan (0.2-1 mg/mL). (D) Flow cytometry analysis of intracellular Alexa Fluor 488 E coli was performed after 30 minutes of culture of cells from healthy controls (open bars) and HIF2A patients (filled bars). (E) Respiratory burst. Neutrophils from healthy controls (open bars) and HIF2A patients (filled bars) were cultured in the presence of dichlorofluorescin only or dichlorofluorescin and N-formyl-Met-Leu-Phe (100 nM) or zymosan A (0.2 mg/ml) and analyzed by flow cytometry. Data show mean and SEM for n = 3.
Neutrophils isolated from patients with gain-of-function HIF2A mutations have enhanced survival but normal function. (A) Apoptosis. Neutrophils from patients with HIF2A mutations (closed squares) or healthy controls (open squares) were cultured for 20 hours with dimethyloxalylglycine (0-100 µM) and apoptosis was determined by morphology. Data are mean ± SEM for n = 3, analyzed by 2-way ANOVA with Bonferroni’s multiple comparison post-test. Overall, there was a significant difference between the control group and the patient group (P < .001), with multiple comparison testing showing statistical significance at the 1 µM concentration (*P < .05). (B) Expression of the HIF targets VEGF, PAI1, and PHD3. TaqMan quantitative PCR analysis of cDNA prepared from freshly isolated neutrophils of patients with HIF2A mutations (filled bars) or healthy controls (open bars). Data are mean and SEM for n = 3. (C-D) Phagocytosis. (C) Phagocytic index was calculated from cytospin slides of neutrophils from healthy controls (open bars) and HIF2A patients (filled bars) prepared after 30 minutes of culture with opsonized zymosan (0.2-1 mg/mL). (D) Flow cytometry analysis of intracellular Alexa Fluor 488 E coli was performed after 30 minutes of culture of cells from healthy controls (open bars) and HIF2A patients (filled bars). (E) Respiratory burst. Neutrophils from healthy controls (open bars) and HIF2A patients (filled bars) were cultured in the presence of dichlorofluorescin only or dichlorofluorescin and N-formyl-Met-Leu-Phe (100 nM) or zymosan A (0.2 mg/ml) and analyzed by flow cytometry. Data show mean and SEM for n = 3.
Overexpression of hif2aa in zebrafish results in delayed resolution of inflammation
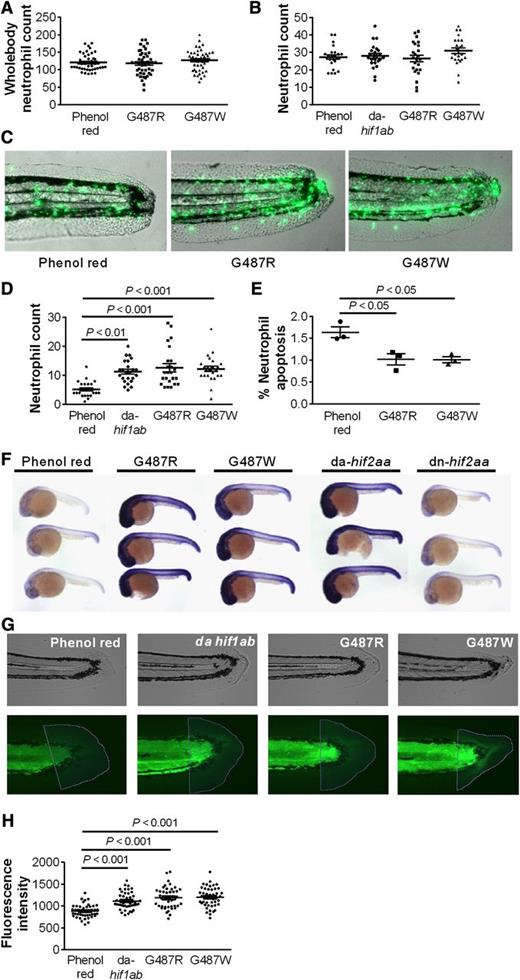
To explore the significance of the reduced neutrophil apoptosis seen in humans with gain-of-function mutations in HIF2A, we mutated the zebrafish ortholog, hif2aa, to produce a protein with an amino acid substitution at the glycine site corresponding to the mutant human protein. We found that replacing the glycine with either arginine (G487R) or tryptophan (G487W), to replicate the HIF2A mutations observed in the patients, did not affect whole fish neutrophil numbers (Figure 3A). In a well-characterized tail injury model of neutrophilic inflammation,23 neutrophil recruitment did not differ between wild-type and mutants (Figure 3B), but the hif2aa-overexpressing fish showed delayed resolution of inflammation (Figure 3C-D). The magnitude of neutrophil persistence was equivalent to that seen with overexpression of hif1ab (Figure 3D) or previously reported with caspase inhibition.23 The increased neutrophil numbers at the site of injury after 24 hours, when inflammation has normally resolved,23 were associated with a significant reduction in neutrophil apoptosis (Figure 3E). Similar results were obtained when we mutated the PHD target sites into nonhydroxylatable amino acids to create a dominant active form of hif2aa (supplemental Figure 2). Importantly, all 3 hif2aa mutants displayed evidence of up-regulation of recognized HIF2A/hif2aa target genes (Figure 3F and data not shown). Injection of a morpholino antisense oligonucleotide to achieve knockdown of the hif2aa binding partner, arnt,30 restored resolution of inflammation in hif2aa overexpressing fish, confirming that the phenotype of delayed inflammation resolution required intact hif2aa signaling (supplemental Figure 2). To study the effects of increased hif2aa expression on wound healing responses, collagen deposition was quantified in the tail transection model, comparing wild type with hif2aa (G487R and G487W) overexpressing and dominant active hif1ab-expressing fish. Both hif2aa and hif1ab overexpressing fish had increased collagen deposition at the injury site 72 hpi (Figure 3G-H) after resolution of the acute neutrophilic response (supplemental Figure 2).
Gain-of-function mutations in the zebrafish HIF2A ortholog hif2aa cause a dominant active-like response in neutrophil behavior and result in increased collagen deposition following zebrafish tailfin transection.hif2aa G487R or G487W RNA (177 pg) or dominant active (da) hif1ab control was injected into 1-cell stage zebrafish mpx:GFP embryos. (A) Whole body total neutrophil numbers at 2 dpf were not altered by injection of hif2aa G487 variants; n = 44 performed as 3 independent experiments. (B-E) Tailfin transection was performed at 2 dpf, and neutrophils were counted at 6 and 24 hours postinjury (hpi). Data shown are mean ± SEM. (B) Injection of dominant active hif2aa variants did not alter the recruitment of neutrophils to the tailfin injury site after 6 hpi. n = 24 performed as 3 independent experiments. (C-D) hif2aa G487 mutations caused a significant increase in neutrophil number at 24 hpi compared with phenol red injected negative controls. (C) Representative overlaid fluorescence and brightfield micrographs (original magnification ×4) imaged on a TE2000U inverted microscope (Nikon, Kingston upon Thames, United Kingdom) at constant exposure. (D) Neutrophil numbers at 24 hpi. n = 24 performed as 3 independent experiments. (E) Injection of hif2aa G487 mutations significantly decreased the percentage of neutrophils at the injury site colabeled with terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling apoptosis staining at 12 hpi. n = 3 performed as independent experiments containing 17 to 36 embryos per injection group per repeat. (F) Evidence of hif2aa target gene activation in zebrafish embryos with overexpressed G487 mutant constructs. Photomicrographs of 24 hpf embryos after injection of dominant active forms of hif2aa RNA (177pg) or dominant negative (dn) hif2aa at the 1-cell stage. Embryos were stained for phd3 expression by in situ hybridization as a representative hif2aa target gene. (G-H) hif2aa G487R or G487W RNA (177pg) or dominant active (da) hif1ab control was injected into 1-cell stage zebrafish mpx:GFP embryos. Tailfins were transected 2 dpf. (G) Representative bright field and fluorescence micrographs (original magnification ×4) of tailfins taken 72 hpi and stained with anti-collagen1 antibody and an anti-mouse Alexa 488 secondary antibody. (H) Mean fluorescence intensity was analyzed using Volocity 5 software. Data represent n = 45 embryos per group performed as 3 independent experiments.
Gain-of-function mutations in the zebrafish HIF2A ortholog hif2aa cause a dominant active-like response in neutrophil behavior and result in increased collagen deposition following zebrafish tailfin transection.hif2aa G487R or G487W RNA (177 pg) or dominant active (da) hif1ab control was injected into 1-cell stage zebrafish mpx:GFP embryos. (A) Whole body total neutrophil numbers at 2 dpf were not altered by injection of hif2aa G487 variants; n = 44 performed as 3 independent experiments. (B-E) Tailfin transection was performed at 2 dpf, and neutrophils were counted at 6 and 24 hours postinjury (hpi). Data shown are mean ± SEM. (B) Injection of dominant active hif2aa variants did not alter the recruitment of neutrophils to the tailfin injury site after 6 hpi. n = 24 performed as 3 independent experiments. (C-D) hif2aa G487 mutations caused a significant increase in neutrophil number at 24 hpi compared with phenol red injected negative controls. (C) Representative overlaid fluorescence and brightfield micrographs (original magnification ×4) imaged on a TE2000U inverted microscope (Nikon, Kingston upon Thames, United Kingdom) at constant exposure. (D) Neutrophil numbers at 24 hpi. n = 24 performed as 3 independent experiments. (E) Injection of hif2aa G487 mutations significantly decreased the percentage of neutrophils at the injury site colabeled with terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling apoptosis staining at 12 hpi. n = 3 performed as independent experiments containing 17 to 36 embryos per injection group per repeat. (F) Evidence of hif2aa target gene activation in zebrafish embryos with overexpressed G487 mutant constructs. Photomicrographs of 24 hpf embryos after injection of dominant active forms of hif2aa RNA (177pg) or dominant negative (dn) hif2aa at the 1-cell stage. Embryos were stained for phd3 expression by in situ hybridization as a representative hif2aa target gene. (G-H) hif2aa G487R or G487W RNA (177pg) or dominant active (da) hif1ab control was injected into 1-cell stage zebrafish mpx:GFP embryos. Tailfins were transected 2 dpf. (G) Representative bright field and fluorescence micrographs (original magnification ×4) of tailfins taken 72 hpi and stained with anti-collagen1 antibody and an anti-mouse Alexa 488 secondary antibody. (H) Mean fluorescence intensity was analyzed using Volocity 5 software. Data represent n = 45 embryos per group performed as 3 independent experiments.
Loss of HIF-2α reduces neutrophilic inflammation and lung injury during inflammation resolution
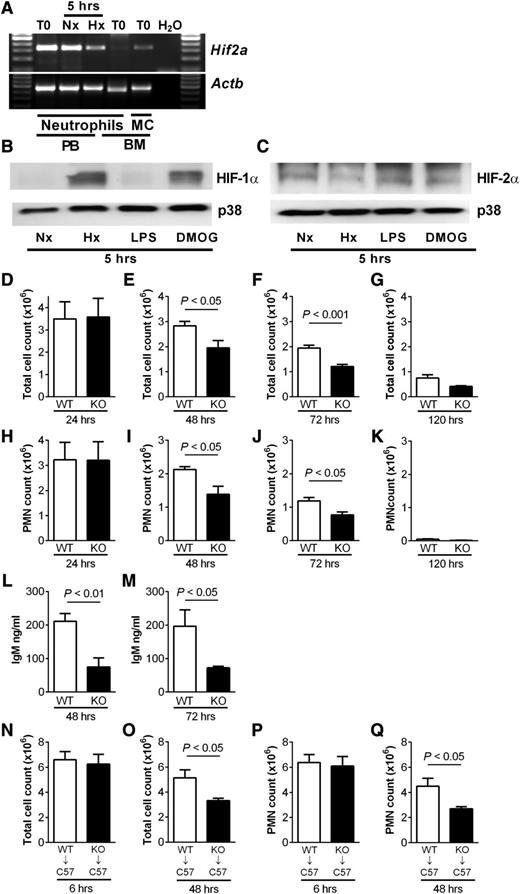
We isolated peripheral blood neutrophils from wild-type mice and confirmed the presence of Hif2a by PCR (Figure 4A). In keeping with previous data, Hif2a was not detected in bone marrow–derived wild-type neutrophils.14 Consistent with our findings in human neutrophils, and in contrast to HIF-1α (Figure 4B), murine neutrophils showed basal expression of HIF-2α protein following normoxic culture, with expression also seen in hypoxia and with LPS stimulation or DMOG treatment (Figure 4C). To investigate the consequences of HIF-2α deficiency, we used mice with myeloid-specific targeted deletion of Hif2a (Hif2aflox/flox;LysMcre+/−).20 HIF-2α–deficient neutrophils showed preserved rates of apoptosis in normoxia and, in contrast to HIF-1α–deficient neutrophils,17 a delay of apoptosis in hypoxia that was equivalent to wild-type cells (supplemental Figure 3). Functional assays on HIF-2α–deficient neutrophils revealed no deficits in respiratory burst, chemotaxis, or phagocytosis, and these cells also showed preserved changes in receptor expression in response to stimulation with LPS (supplemental Figure 4).
Reduced neutrophilic inflammation and lung injury during inflammation resolution in mice with myeloid cell deficiency of HIF-2α (A) Expression profile of Hif2a in murine peripheral blood and bone marrow neutrophils. Neutrophils isolated by magnetic negative selection from peripheral blood (PB) or bone marrow (BM) or bone marrow mononuclear cells (MCs) were cultured in normoxia (Nx) or hypoxia (Hx) for 5 hours or lysed when freshly isolated (T0). cDNA was amplified using custom Hif2a primers, and a representative gel image of n = 3 is shown. (B-C) Murine peripheral blood neutrophils express (B) HIF-1α and (C) HIF-2α. Representative western blots of lysates from neutrophils cultured in normoxia (Nx) or hypoxia (Hx) or stimulated in normoxia with LPS (10 ng/mL) or DMOG (100 µM) for 5 hours. (D-M) Hif2aflox/flox;LysMCre−/− controls (WT) and littermate Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS (3 mg). BAL was performed at 24, 48, 72, and 120 hours. (D-G) Total cell counts were determined by hemocytometer and (H-K) neutrophil counts by cytospin analysis. (L-M) IgM levels were determined in BAL fluid by enzyme-linked immunosorbent assay. Data are mean and SEM for controls (open bars) and HIF-2α–deficient mice (filled bars), n = 5. (N-Q) Wild-type (WT) C57BL/6 mice (C57) were irradiated (12 fractions of 1Gy), injected with bone marrow from Hif2aflox/flox;LysMCre−/− (WT) or Hif2aflox/flox;LysMCre+/− (KO) mice, and allowed to reconstitute for 5 weeks before challenge with nebulized LPS. Bronchoalveolar lavage was performed at 6 and 48 hours. (N-O) Total cell counts determined by hemocytometer and (P-Q) neutrophil counts determined by cytospin. Data are mean and SEM for WT→C57 (open bars) and KO→C57 mice (filled bars), minute n = 5.
Reduced neutrophilic inflammation and lung injury during inflammation resolution in mice with myeloid cell deficiency of HIF-2α (A) Expression profile of Hif2a in murine peripheral blood and bone marrow neutrophils. Neutrophils isolated by magnetic negative selection from peripheral blood (PB) or bone marrow (BM) or bone marrow mononuclear cells (MCs) were cultured in normoxia (Nx) or hypoxia (Hx) for 5 hours or lysed when freshly isolated (T0). cDNA was amplified using custom Hif2a primers, and a representative gel image of n = 3 is shown. (B-C) Murine peripheral blood neutrophils express (B) HIF-1α and (C) HIF-2α. Representative western blots of lysates from neutrophils cultured in normoxia (Nx) or hypoxia (Hx) or stimulated in normoxia with LPS (10 ng/mL) or DMOG (100 µM) for 5 hours. (D-M) Hif2aflox/flox;LysMCre−/− controls (WT) and littermate Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS (3 mg). BAL was performed at 24, 48, 72, and 120 hours. (D-G) Total cell counts were determined by hemocytometer and (H-K) neutrophil counts by cytospin analysis. (L-M) IgM levels were determined in BAL fluid by enzyme-linked immunosorbent assay. Data are mean and SEM for controls (open bars) and HIF-2α–deficient mice (filled bars), n = 5. (N-Q) Wild-type (WT) C57BL/6 mice (C57) were irradiated (12 fractions of 1Gy), injected with bone marrow from Hif2aflox/flox;LysMCre−/− (WT) or Hif2aflox/flox;LysMCre+/− (KO) mice, and allowed to reconstitute for 5 weeks before challenge with nebulized LPS. Bronchoalveolar lavage was performed at 6 and 48 hours. (N-O) Total cell counts determined by hemocytometer and (P-Q) neutrophil counts determined by cytospin. Data are mean and SEM for WT→C57 (open bars) and KO→C57 mice (filled bars), minute n = 5.
A neutrophil-mediated LPS-induced model of acute lung injury was used to determine the consequences of HIF-2α deficiency in vivo. Mice with myeloid-specific deletion of Hif2a had very few neutrophils in unstimulated airways (supplemental Figure 5) and displayed normal recruitment of neutrophils to the lung (Figure 4D,H), but thereafter, had significantly lower BAL total cell counts and neutrophil counts than controls at 48 (Figure 4E,I) and 72 hours (Figure 4F,J), and neutrophil clearance by 120 hours (Figure 4K). Importantly, the lower neutrophil counts were accompanied by a significant reduction in lung injury in the HIF-2α–deficient mice, as determined by IgM release (Figure 4L-M). Cytokine and chemokine profiles in plasma and BAL fluid showed no differences between strains, in keeping with their equivalent neutrophil recruitment, and there was no difference in macrophage efferocytosis (supplemental Figure 5). To exclude a role for alveolar macrophages in determining the outcome of the neutrophilic response, experiments were repeated in wild-type C57BL/6 mice following a fractional radiation protocol28 and bone marrow reconstitution with either Hif2aflox/flox;LysMcre+/− or Hif2aflox/flox;LysMcre−/− cells (supplemental Figure 7). Neutrophil recruitment did not differ between the groups, but significant reductions in total cell and neutrophil counts were again observed at 48 hours (Figure 4N-Q).
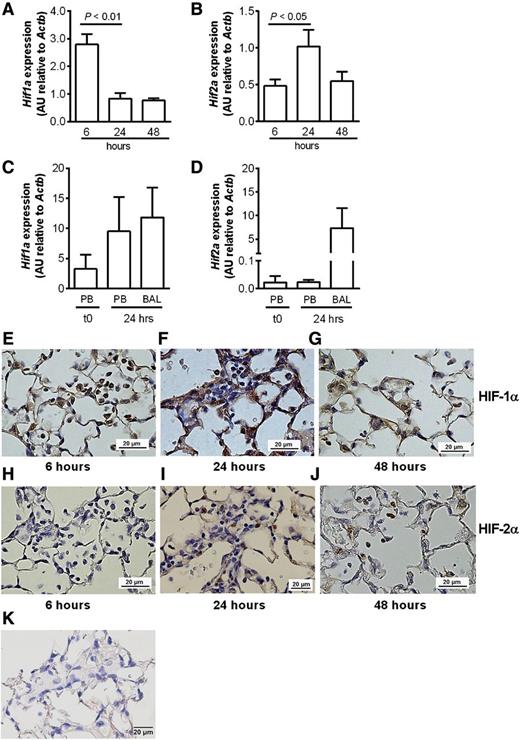
Temporal and regional regulation of HIF-1α and HIF-2α transcript and protein expression in neutrophils following acute lung injury
To compare tissue-recruited and circulating neutrophil expression profiles of Hif1a and Hif2a, neutrophils were isolated from the blood and lungs of animals challenged with intrapulmonary LPS. Differential regulation of Hif1a and Hif2a was observed in wild-type LPS-treated animals, with down-regulation of Hif1a mRNA in BAL samples over time following LPS challenge (Figure 5A). In marked contrast, Hif2a mRNA expression was maintained during the resolution phase of acute lung injury (Figure 5B). There was also selective induction of Hif2a (Figure 5D) but not Hif1a (Figure 5C) mRNA in cells harvested from the BAL fluid at 24 hours (91 ± 1.7% neutrophils, n = 3) compared with paired samples of circulating neutrophils and peripheral blood neutrophils of unchallenged mice, and this was validated in flow sorted bronchoalveolar neutrophils (data not shown). Immunohistochemistry confirmed the changes in Hif transcripts correlated with expression of HIF proteins in recruited neutrophils, with early expression of HIF-1α but not HIF-2α (Figure 5E,H) and delayed and persistent expression of HIF-2α (Figure 5I,J) accompanied by reduced HIF-1 α expression between the 24- and 48-hour time points (Figure 5F-G). No HIF-2α staining was observed in myeloid cells of HIF-2α–deficient mice (Figure 5K).
Differential regulation of HIF1 and HIF2 in circulating neutrophils and neutrophils recruited to the lungs following LPS-induced lung injury. (A-D) C57BL/6 mice were instilled with intratracheal LPS (0.3 µg). BAL was performed at 6, 24, and 48 hours. (A) Hif1a and (B) Hif2a expression in BAL cell lysates from C57BL/6 mice determined by Taqman and normalized to Actb. (C) Hif1a and (D) Hif2a expression in freshly isolated (t0) peripheral blood neutrophils (PBs), or peripheral blood neutrophils and BAL cells isolated 24 hours after LPS instillation. Data are mean and SEM for n = 3. (E-J) Immunohistochemistry showing expression of (E-G) HIF-1α and (H-J) HIF-2α in neutrophils of WT mice (E,H) 6, (F,I) 24, and (G,J) 48 hours following challenge with nebulized LPS (3 mg). (K) Immunohistochemistry showing no HIF-2α expression in myeloid-specific HIF-2α–deficient mouse lungs. Original magnification ×400. Images were taken using a Nikon Eclipse E600 microscope, with a Nikon DS-Ri1 camera, and processed with NIS-Elements Basic Research software (Nikon, Kingston upon Thames, United Kingdom).
Differential regulation of HIF1 and HIF2 in circulating neutrophils and neutrophils recruited to the lungs following LPS-induced lung injury. (A-D) C57BL/6 mice were instilled with intratracheal LPS (0.3 µg). BAL was performed at 6, 24, and 48 hours. (A) Hif1a and (B) Hif2a expression in BAL cell lysates from C57BL/6 mice determined by Taqman and normalized to Actb. (C) Hif1a and (D) Hif2a expression in freshly isolated (t0) peripheral blood neutrophils (PBs), or peripheral blood neutrophils and BAL cells isolated 24 hours after LPS instillation. Data are mean and SEM for n = 3. (E-J) Immunohistochemistry showing expression of (E-G) HIF-1α and (H-J) HIF-2α in neutrophils of WT mice (E,H) 6, (F,I) 24, and (G,J) 48 hours following challenge with nebulized LPS (3 mg). (K) Immunohistochemistry showing no HIF-2α expression in myeloid-specific HIF-2α–deficient mouse lungs. Original magnification ×400. Images were taken using a Nikon Eclipse E600 microscope, with a Nikon DS-Ri1 camera, and processed with NIS-Elements Basic Research software (Nikon, Kingston upon Thames, United Kingdom).
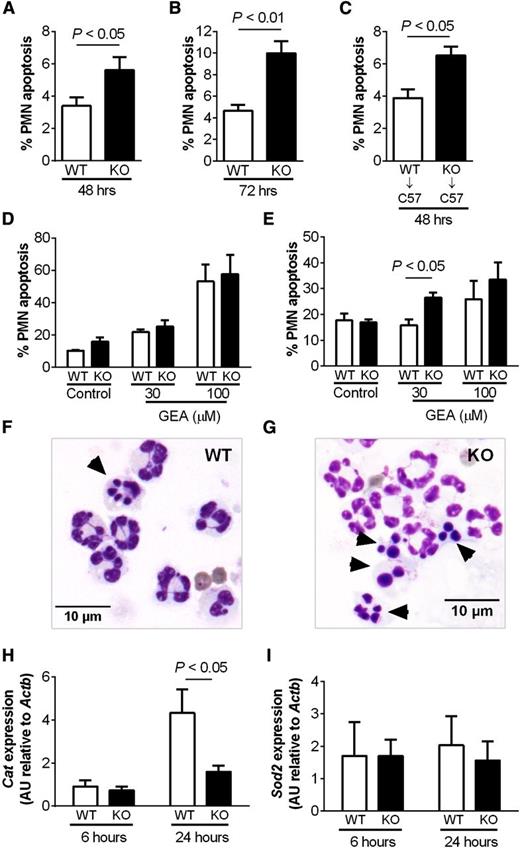
HIF-2α–deficient inflammatory neutrophils display an intrinsic apoptosis phenotype and increased sensitivity to nitrosative stress
Apoptosis of inflammatory neutrophils recruited to the airways following LPS challenge was quantified using morphologic criteria, with subsequent verification by annexinV/To-Pro-3 positivity (supplemental Figure 5I-J). Increased neutrophil apoptosis was observed in the HIF-2α–deficient mice at 48 and 72 hours following LPS challenge (Figure 6A-B), and this was recapitulated in the adoptive transfer experiments where C57BL/6 mice were reconstituted with bone marrow from Hif2aflox/flox;LysMcre+/−mice (Figure 6C). When studied, ex vivo inflammatory neutrophils, in contrast to peripheral blood neutrophils, displayed increased rates of apoptosis following in vitro culture with the NO donor GEA3162 (Figure 6D-G). This correlated with a significant reduction in transcript abundance of the antioxidant catalase (Cat), a transcript induced in human neutrophils following hypoxic culture (data not shown), but not Sod2 (Figure 6H-I). Expression of key neutrophil apoptosis regulators Bclxl, Siva1, Mcl1, and Nfkb did not differ between wild-type and HIF-2α–deficient neutrophils (supplemental Figure 6).
Increased apoptosis of HIF-2α–deficient inflammatory neutrophils. (A-B) Hif2aflox/flox;LysMCre−/− (WT) and littermate Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS (3 mg). BAL was performed at 48 and 72 hours. Neutrophil apoptosis was determined by morphology. Data are mean and SEM for n = 5. (C) Wild-type (WT) C57BL/6 mice (C57) were irradiated (12 fractions of 1Gy), injected with bone marrow from Hif2aflox/flox;LysMCre−/− (WT) or Hif2aflox/flox;LysMCre+/− (KO) mice, and allowed to reconstitute for 5 weeks before challenge with nebulized LPS. BAL was performed at 48 hours, and neutrophil apoptosis was determined by morphology. Data are mean and SEM for min n = 5. (D-G) Hif2aflox/flox;LysMCre−/− (WT) and Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS. At 24 hours, peripheral blood neutrophils isolated by (D) negative magnetic selection and (E) inflammatory neutrophils recovered from BAL were cultured with GEA3162 (GEA) for 9 hours. Apoptosis was determined by morphology. Data are mean and SEM for n = 4. (F-G) Representative cytospin images showing (F) WT and (G) KO inflammatory neutrophils recovered from BAL following 9 hours of culture with 30 µM GEA3162. Original magnification ×1000. (H-I) WT and KO mice were challenged with nebulized LPS (3 mg). BAL was performed at 6 and 24 hours, and expression of catalase (Cat) and superoxide dismutase 2 (Sod2) in BAL cell lysates was determined by Taqman and normalized to Actb. Data are mean and SEM for minute n = 4.
Increased apoptosis of HIF-2α–deficient inflammatory neutrophils. (A-B) Hif2aflox/flox;LysMCre−/− (WT) and littermate Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS (3 mg). BAL was performed at 48 and 72 hours. Neutrophil apoptosis was determined by morphology. Data are mean and SEM for n = 5. (C) Wild-type (WT) C57BL/6 mice (C57) were irradiated (12 fractions of 1Gy), injected with bone marrow from Hif2aflox/flox;LysMCre−/− (WT) or Hif2aflox/flox;LysMCre+/− (KO) mice, and allowed to reconstitute for 5 weeks before challenge with nebulized LPS. BAL was performed at 48 hours, and neutrophil apoptosis was determined by morphology. Data are mean and SEM for min n = 5. (D-G) Hif2aflox/flox;LysMCre−/− (WT) and Hif2aflox/flox;LysMCre+/− (KO) mice were challenged with nebulized LPS. At 24 hours, peripheral blood neutrophils isolated by (D) negative magnetic selection and (E) inflammatory neutrophils recovered from BAL were cultured with GEA3162 (GEA) for 9 hours. Apoptosis was determined by morphology. Data are mean and SEM for n = 4. (F-G) Representative cytospin images showing (F) WT and (G) KO inflammatory neutrophils recovered from BAL following 9 hours of culture with 30 µM GEA3162. Original magnification ×1000. (H-I) WT and KO mice were challenged with nebulized LPS (3 mg). BAL was performed at 6 and 24 hours, and expression of catalase (Cat) and superoxide dismutase 2 (Sod2) in BAL cell lysates was determined by Taqman and normalized to Actb. Data are mean and SEM for minute n = 4.
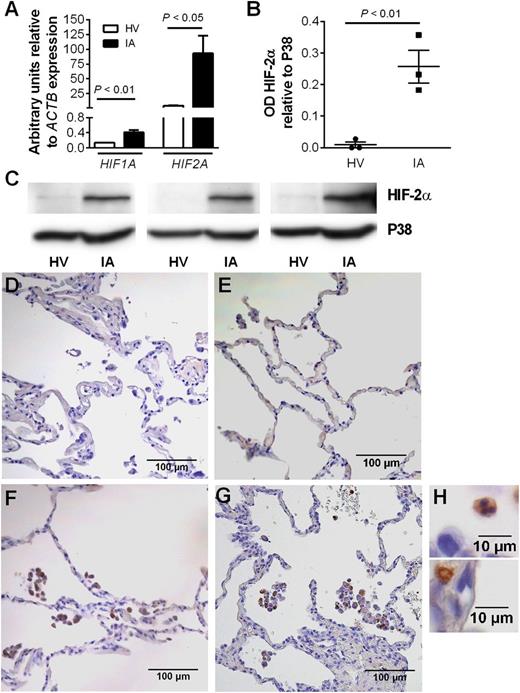
Neutrophils from patients with chronic inflammatory disease show enhanced HIF-2α expression
Peripheral blood neutrophils isolated from patients with inflammatory arthritis displayed enhanced HIF1A and HIF2A mRNA expression (Figure 7A). HIF-2α protein expression was also significantly higher in circulating neutrophils of arthritis patients than of healthy controls (Figure 7B-C). Interestingly, this was selective for HIF-2α, with no increase in HIF-1α protein observed (data not shown). Neutrophils recruited to the airways in both mild and severe COPD also displayed strong HIF-2α staining, in marked contrast to epithelium where no HIF-2α expression was demonstrated (Figure 7D,H).
Expression of HIF-2α is up-regulated in neutrophils from patients with active inflammatory arthritis and is seen in neutrophils within lung biopsies from patients with COPD. (A) Expression of HIF1A and HIF2A in inflammatory arthritis patients (filled bars) and controls (open bars) was determined by TaqMan analysis of cDNA from freshly isolated peripheral blood neutrophils with data normalized to ACTB expression. Data are mean and SEM for n = 4. (B-C) HIF-2α protein expression is significantly higher in neutrophils from patients with inflammatory arthritis (IA) than healthy volunteers (HV). Freshly isolated neutrophils were lysed and proteins separated by SDS-PAGE. Blots were probed for HIF-2α, and densitometry data were normalized to p38 MAPK. (B) Data are mean and SEM for n = 3. (C) HIF-2α blots of circulating neutrophil lysates from 3 healthy volunteers (HV) and 3 patients with inflammatory arthritis (IA). (D-H) Immunohistochemistry showing HIF-2α expression in lung biopsies from a (E) nonsmoker and patients with (F) mild or (G-H) severe COPD. Images are representative of n = 2. D is a section stained with an isotype control. Original magnification: (D-G) ×200; (H) ×1000.
Expression of HIF-2α is up-regulated in neutrophils from patients with active inflammatory arthritis and is seen in neutrophils within lung biopsies from patients with COPD. (A) Expression of HIF1A and HIF2A in inflammatory arthritis patients (filled bars) and controls (open bars) was determined by TaqMan analysis of cDNA from freshly isolated peripheral blood neutrophils with data normalized to ACTB expression. Data are mean and SEM for n = 4. (B-C) HIF-2α protein expression is significantly higher in neutrophils from patients with inflammatory arthritis (IA) than healthy volunteers (HV). Freshly isolated neutrophils were lysed and proteins separated by SDS-PAGE. Blots were probed for HIF-2α, and densitometry data were normalized to p38 MAPK. (B) Data are mean and SEM for n = 3. (C) HIF-2α blots of circulating neutrophil lysates from 3 healthy volunteers (HV) and 3 patients with inflammatory arthritis (IA). (D-H) Immunohistochemistry showing HIF-2α expression in lung biopsies from a (E) nonsmoker and patients with (F) mild or (G-H) severe COPD. Images are representative of n = 2. D is a section stained with an isotype control. Original magnification: (D-G) ×200; (H) ×1000.
Discussion
Innate immune cells must function competently in the hypoxic microenvironment of infected and inflamed tissues. Oxygen-sensing transcription factors, notably HIF, allow them to adapt to these conditions. HIF-1α has fundamental roles in cellular oxygen sensing, and metabolic adaptation to hypoxia in many tissues is essential for neutrophil survival in hypoxia and regulates myeloid cell bacterial killing.6,13,16 Myeloid-specific HIF-1α deficiency reduces inflammation,but increases susceptibility to bacterial infections both locally and systemically.13,16 This infection risk limits the utility of direct targeting of HIF-1α as an anti-inflammatory approach, hence our search for more selective regulators of neutrophil function and fate. With recent evidence suggesting a role for HIF-2α in myeloid inflammatory responses,14 and a number of unique functions already ascribed to both HIF-1α and HIF-2α, we proposed HIF-2α would have a more selective role in neutrophil biology and the regulation of inflammation resolution.
In keeping with previous findings from bone marrow–derived neutrophils and murine neutrophil cell lines,14 we were unable to detect HIF-2α mRNA or protein in immature murine bone marrow neutrophils. Althoughe we were able to detect low levels of HIF2A mRNA in highly pure peripheral blood neutrophils, higher levels of HIF-2 mRNA were seen following neutrophil activation in the context either of a systemic inflammatory condition (arthritis) or recruitment to an injured site (LPS-mediated acute lung injury). Transcriptional regulation of HIF mRNA expression has been described by Takeda et al,20 who showed HIF1A and HIF2A mRNA are differentially expressed in M1- and M2-polarized macrophages, with labile HIF1A mRNA displaying a relatively short half-life and conversely HIF2A mRNA being much more stable with a lower rate of turnover. Of note, these changes in mRNA expression occurred independent of oxygen availability and were themselves a strong predictor of protein abundance. In keeping with this, we also observed much higher levels of HIF-2α protein in freshly isolated neutrophils from patients with inflammatory arthritis compared with healthy control neutrophils and in neutrophils recruited to the lung both in patients with mild and severe COPD and in a murine acute lung injury model.
The importance of HIF-2α accumulation with respect to disease outcomes is unknown but, through access to 3 patients with known gain-of-function mutations in the HIF2A gene, we were able to assess the functional consequences of HIF-2α overexpression in neutrophils. Baseline rates of constitutive apoptosis were consistently lower in patient neutrophils than controls, although further reductions in apoptosis were achieved by stimulation with DMOG. This is consistent with experimental evidence showing only partial activation of HIF-2α in these patients, so that hydroxylase inhibition could further stabilize HIF-2α and stabilize HIF-1α.29 Neutrophils derived from patients with gain-of-function mutations in HIF2A also showed increased HIF-2α target gene expression (VEGF, PAI1, and PHD3) to a level equivalent to that previously described for hypoxic culture.17
Delayed neutrophil apoptosis has been implicated in persistence of inflammation in animal models,5,31 but whether the increase in HIF-2α expression and associated intrinsic delay of neutrophil apoptosis might predispose these patients to inflammatory disease is not known, not least because of the rarity of the condition and the dominant clinical phenotype of erythrocytosis and its consequences. To directly address the importance of selective HIF-2α stabilization for regulation of neutrophil survival and resolution of inflammation, we replicated the human gain-of-function HIF2A mutation in the genetically tractable zebrafish.23,25 The hydroxylation sites of HIF-α subunits are highly conserved across species,25 with an overall amino acid homology of 58% between zebrafish hif2aa and human HIF-2α. Using mutated zebrafish RNA, we expressed hif2aa mutated at the same glycine residues as are altered in the patients with gain-of-function HIF2A mutations. Expression of mutant hif2aa led to impaired resolution of inflammation in a tail injury model that was equivalent to overexpression of dominant active hif1ab and of a similar magnitude to previous results using the pan-caspase inhibitor, zVD.fmk.23 These findings highlight that in vivo hif2aa can modulate neutrophil survival to the same order of magnitude as hif1ab. Importantly, the impairment in inflammation resolution also has consequences for wound healing responses, because overexpression of hif2aa results in localized increases in collagen deposition 72 hpi.
To address the therapeutic potential of selectively targeting HIF-2α, we investigated the consequences of HIF-2α deficiency using mice with myeloid-specific deletion of Hif2a, as previously described.20 In marked contrast to HIF-1α deficiency,16 we found no impairment of neutrophil chemotaxis, phagocytosis, or respiratory burst in HIF-2α–deficient neutrophils. In an in vivo model of LPS-mediated lung injury, HIF-2α deficiency was associated with reduced neutrophilic inflammation during resolution, with fewer neutrophils in BAL samples, an increase in neutrophil apoptosis, and a reduction in lung damage and vascular leak. Given that the changes in neutrophil accumulation could in part be a consequence of altered macrophage function, the lung injury model was repeated in wild-type mice following fractional radiation, which both preserves the recipient alveolar macrophage population and epithelium and enables bone marrow reconstitution.28 Reduced neutrophilic inflammation was again observed in the context of marrow reconstitution with Hif2aflox/flox;LysMcre+/− cells. An intrinsic neutrophil phenotype dependent on HIF-2α expression was subsequently confirmed through the ex vivo study of inflammatory neutrophils isolated from the airways following LPS challenge. In this setting, HIF-2α–deficient neutrophils were shown to have increased sensitivity to nitrosative stress-induced apoptosis, matched with a significant reduction in catalase transcript abundance, previously identified as a Hif2a-regulated gene.32 Together, these data suggest that during inflammation resolution, the neutrophilic inflammatory process is regulated by HIF-2α rather than HIF-1α expression through the intrinsic regulation of neutrophil apoptosis. This is further supported by HIF-1α and HIF-2α expression data. First, we observed that neutrophils recruited from the circulation to the lung following LPS challenge significantly up-regulate Hif2a mRNA but not Hif1a mRNA. Second, Hif2a mRNA expression persists in BAL cells during inflammation resolution after Hif1a mRNA expression has substantially reduced.
The phenotype of reduced neutrophilic inflammation during resolution of an acute lung injury contrasts with our findings in PHD3-deficient animals, in which we observed a specific role for PHD3 in regulating neutrophilic inflammation in the context both of whole animal and localized tissue hypoxia.17 Although PHD3 is essential for neutrophil survival and inflammatory responses in hypoxia, we propose that the effects of HIF-2α deficiency in inflammatory neutrophils occur independently of oxygen tension, and targeting HIF-2α may therefore be of greater clinical utility in inflammation in tissues, such as the lung, where oxygen tension can vary widely.33 Through modulation of tumor cell apoptosis, HIF-2α has emerged as a potential therapeutic target in cancer biology.34,35 Our data now implicate HIF-2α in neutrophilic inflammation and propose that selective inhibition of HIF-2α may allow effective control of neutrophil-mediated inflammation and of inflammation resolution without compromising host defenses.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Rachael Kilding, Dr Mohammed Akil, and Dr John Boulton for help recruiting patients with inflammatory arthritis and performing synovial fluid aspiration; Emma Connelly and Yvonne Stephenson for help with western blots and immunohistochemistry; Dr Andrew Cowburn for help with in vivo murine models of regional neutrophilic inflammation; Dr Colin Bingle for his help obtaining lung biopsy specimens; and Prof Matthias Mack for useful discussion of in vivo murine models.
This work was supported by a Medical Research Council (MRC) Clinical Training Fellowship award to A.A.R.T. (G0802255), a Wellcome Trust Intermediate Clinical Fellowship award to S.R.W. (078244), a Wellcome Trust Senior Clinical Fellowship award to S.R.W. (098516), a Wellcome Trust Senior Clinical Fellowship award to D.H.D. (076945), a MRC Senior Clinical Fellowship award to S.A.R. (G0701932), and a British Lung Foundation fellowship to H.M.M. (F05/7). The National Institute for Health Research Sheffield Biomedical Research Unit in Cardiovascular Disease and zebrafish work were supported by an MRC Centre Grant (G0700091).
Authorship
Contribution: A.A.R.T. performed the research and wrote the manuscript; P.M.E., H.M.M., S.E., K.R.H., A.L., L.W., S.P., G.S., and E.E.M. performed the research; F.F., F.J.V.E., V.L.K., C.W.P., P.A.R., M.J.P., M.C.S., R.S.J., and S.A.R. provided access to novel analytical tools, transgenic organisms, and patient samples; S.A.R., I.S., D.H.D., and E.R.C. interpreted data; and M.K.B.W. and S.R.W. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: C.W.P. is a scientific cofounder of, and holds equity in, ReOx Ltd. The remaining authors declare no competing financial interests.
Correspondence: Sarah Walmsley, Academic Unit of Respiratory Medicine, The Medical School, University of Sheffield, Sheffield S10 2RX, United Kingdom; e-mail: s.walmsley@sheffield.ac.uk; and Moira Whyte, Academic Unit of Respiratory Medicine, The Medical School, University of Sheffield, Sheffield S10 2RX, United Kingdom; e-mail: m.k.whyte@sheffield.ac.uk.
References
Author notes
M.K.B.W. and S.R.W. contributed equally to the work.