Key Points
I-BET151 and I-BET-762 induce cell cycle arrest and apoptosis in myeloma cells associated with MYC downregulation and HEXIM1 upregulation.
Preclinical functional and pharmacologic profiling of I-BET762 supports its use in phase 1 clinical studies.
Abstract
The bromodomain and extraterminal (BET) protein BRD2-4 inhibitors hold therapeutic promise in preclinical models of hematologic malignancies. However, translation of these data to molecules suitable for clinical development has yet to be accomplished. Herein we expand the mechanistic understanding of BET inhibitors in multiple myeloma by using the chemical probe molecule I-BET151. I-BET151 induces apoptosis and exerts strong antiproliferative effect in vitro and in vivo. This is associated with contrasting effects on oncogenic MYC and HEXIM1, an inhibitor of the transcriptional activator P-TEFb. I-BET151 causes transcriptional repression of MYC and MYC-dependent programs by abrogating recruitment to the chromatin of the P-TEFb component CDK9 in a BRD2-4–dependent manner. In contrast, transcriptional upregulation of HEXIM1 is BRD2-4 independent. Finally, preclinical studies show that I-BET762 has a favorable pharmacologic profile as an oral agent and that it inhibits myeloma cell proliferation, resulting in survival advantage in a systemic myeloma xenograft model. These data provide a strong rationale for extending the clinical testing of the novel antimyeloma agent I-BET762 and reveal insights into biologic pathways required for myeloma cell proliferation.
Introduction
Multiple myeloma is a malignancy of plasma cells, the terminally differentiated, immunoglobulin-secreting B cells.1 The primary tumor-initiating genetic events include translocations and hyperdiploidy, while secondary events such as activation of the oncogene MYC drive disease progression.2,3 Despite recent therapeutic advances and improved survival, myeloma remains an incurable malignancy, and nearly all patients eventually succumb to treatment-refractory disease, which is often highly proliferative.4
Recently, inhibitors have been developed that target the acetyl-binding pockets of the bromodomains of the bromodomain and extraterminal domain (BET) family of proteins BRD2-4 and BRDT. BET proteins activate transcription through their ability to bind to acetyl-modified lysine residues of histone tails5,6 thereby serving as chromatin scaffolds that recruit the P-TEFb and PAFc1 complexes to RNA polymerase II (RNA Pol II), thus ensuring transcriptional initiation and elongation.7-11 Two classes of BET inhibitors, benzodiazepines and quinolines, have been shown to have significant antiproliferative activity against a variety of hematologic tumors.12 Specifically, the benzodiazepine JQ113 was shown to be active against myeloma,14 lymphoma,15 and acute lymphoblastic leukemia16 in vitro and in vivo; of the quinoline class of BET protein inhibitors, I-BET151 was shown to have preclinical activity against acute leukemia, including mixed lineage leukemia–related acute myeloid leukemia.10,11 At the mechanistic level, a unifying theme for both JQ1 and I-BET151 is their ability to inhibit transcription of MYC and MYC-dependent oncogenic programs.11,14,15 Oncogenic MYC activation and overexpression is common in myeloma17,18 and can come about by a variety of genetic mechanisms, including chromosomal rearrangements that bring MYC transcriptional regulation under the influence of immunoglobulin gene regulatory areas, gene amplification, or by oncogenic RAS-driven MYC overexpression.2,19
Herein, by combining in vitro and in vivo approaches with global transcriptomics, we identify genes and biologic processes that underpin the antimyeloma activity of the tool molecule I-BET151. In addition, we translate this knowledge to the structurally distinct clinical compound I-BET762,20 which for the first time demonstrates that it displays significant antitumor activity in vivo in an animal model of systemic myeloma.
Materials and methods
Primary myeloma cells
Primary myeloma CD138+ plasma cells were isolated in high purity (>98%; data not shown) from bone marrow (BM) samples by using CD138 microbeads and magnetic cell sorting (Miltenyi Biotec). The samples were taken at diagnosis or relapse of the disease as part of the patients’ clinical assessment at the Hammersmith Hospital after appropriate written informed consent was obtained in accordance with the Declaration of Helsinki. The study was approved by the local research ethics committee.
Cell culture and inhibitors
For in vitro cell proliferation and apoptosis assays, myeloma cell lines were cultured by using RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich), 2 mM l-glutamine, penicillin 500 IU/mL, and streptomycin 500 μg/mL. Cells were placed in 96-well U-bottom plates at final concentration of 0.2 × 106 cells per milliliter in a humidified incubator with 5% CO2 at 37°C. For stroma vs nonstroma experiments, myeloma cells were placed in flat-bottom 96-well plates with MS5 cells at >90% confluence or in wells without stroma. Compounds (ie, I-BET151, I-BET762, the inactive isomer I-BET768, and JQ1) were serially diluted into media and added to the cultures at the indicated concentrations, starting from a 10-mM dimethylsulfoxide (DMSO) stock solution.
Primary myeloma cells were cultured in flat-bottom 96-well plates in the presence of MS5 stroma cells by using complete medium as above, supplemented with interleukin-6 (IL-6) at 5 ng/mL.
Gene expression profiling
Total RNA was extracted from H929 and KMS12BM cells after culture with I-BET151 1 μM or vehicle (DMSO) for 6 hours (3 independent experiments) by using the RNeasy Mini Kit (Qiagen). Genomic DNA contamination was eliminated by using columns (Qiagen) and deoxyribonuclease digestion. The extracted RNA was quantified in a Nanodrop spectrophotometer, while purity and integrity were confirmed on an Agilent 2100 Bioanalyser with RNA Nano Chips. Complementary DNA was produced from 150 ng total RNA input, using the Ambion WT Expression Kit. The complementary DNA was then fragmented and labeled by using the GeneChip WT Terminal Labeling Kit (Affymetrix) and hybridized on Human Gene ST1.0 Arrays (Affymetrix) on a GeneChip Fluidics Station 450. The arrays were scanned in a GeneChip Scanner 3000 7G with autoloader.
For gene set enrichment analysis (GSEA), core probes of Affymetrix HuGene_1_0-st array were used to summarize expressions at the transcript level. Robust multi-array average21 was applied for background correction using the antigenomic probes and quantile normalization. R and Bioconductor package xps were used for data extraction.
GSEA v2 and the gene sets categories of C2 and C3 v3.0 were downloaded from MSigDB v3.0.22 Gene expression was fed from normalized and summarized core probes at the transcript level. Gene sets with nominal P value < .01 and false discovery rate (FDR) q < .05 were chosen as significant for downstream analysis. The gene expression profiling (GEP) data are available online in ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-2122.
Xenotransplantation experiments
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were bred and maintained in-house at Imperial College in accordance with the 1986 Animal Scientific Procedures Act and under a United Kingdom Government Home Office–approved project license. In total, 5 × 106 KMS11 myeloma cells were injected subcutaneously into 9- to 12-week-old NSG mice. When tumors were ≥5 mm in maximum diameter, mice were randomized to receive once daily intraperitoneal injection of either I-BET151 30 mg/kg in 0.9% NaCl plus Kleptose hydroxypropyl betadex 10% (w/v) and DMSO 5% (v/v) pH 5.0 or vehicle solution for a maximum of 21 days.
The antimyeloma efficacy of orally administered I-BET762 was tested in a systemic xenograft myeloma model. For this purpose, sublethally irradiated (200 cGy) NOD/SCID mice age 9 to 11 weeks were given 107 OPM-2 myeloma cells via tail vein injection. On day 15 following inoculation, animals were started on oral treatment with I-BET762 at escalating doses or vehicle (1% methylcellulose and 0.2% sodium lauryl sulfate), which was continued up to day 83. Specifically, we treated 1 group of mice with vehicle and 4 groups with different dosing schedules of I-BET762: 3 mg/kg per day; 10 mg/kg per day; 30 mg/kg on alternate days; and 30 to 20 mg/kg per day (ie, 30 mg/kg per day for 14 days, followed by 2 weeks [days 15 to 31] off treatment [drug was withheld due to a decline in body weight until animals had regained weight], followed by 20 mg/kg per day until termination of the experiment [days 43 to 82]). Blood samples (∼70 μL) were removed at 0.5 hours after oral administration of I-BET762 on day 15 (treatment initiation); days 27, 45, and 82 (3, 10, and 20 to 30 mg/kg once per day groups only); and day 83 (30 mg/kg once every other day group only). The blood was centrifuged to obtain 20 μL plasma and stored at −20°C prior to analysis for I-BET762 by using a specific liquid chromatography/mass spectrometry/mass spectrometry assay.
Serum human λ light chain (hLC) was measured with enzyme-linked immunosorbent assay, and the frequency of BM CD38+ human myeloma cells was measured by flow cytometry and by histologic examination (in euthanized animals). All studies were conducted in accordance with the GlaxoSmithKline Policy on the Care, Welfare and Treatment of Laboratory Animals and were reviewed by the Institutional Animal Care and Use Committee either at GlaxoSmithKline or by the ethical review process at the institution where the work was performed.
Statistical analysis
Statistical analysis was performed by using GraphPad Prism 5 software. Kaplan-Meier curve analysis was used to determine survival and probability of tumor size doubling, with groups compared by using the log-rank test. Paired data were tested by using Wilcoxon rank test. P values <.05 were deemed statistically significant.
Cell proliferation assays, flow cytometry, cell cycle analysis, apoptosis assays, chromatin immunoprecipitation analysis, enzyme-linked immunosorbent assay, gene expression studies by real time quantitative polymerase chain reaction (RQ-PCR), differential salt extraction, western blotting, and histopathology are detailed in the supplemental data on the Blood Web site.
Results
I-BET151 induces cell cycle arrest and apoptosis in myeloma cell lines
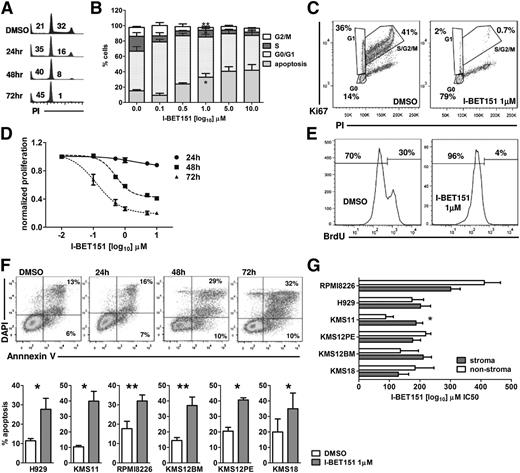
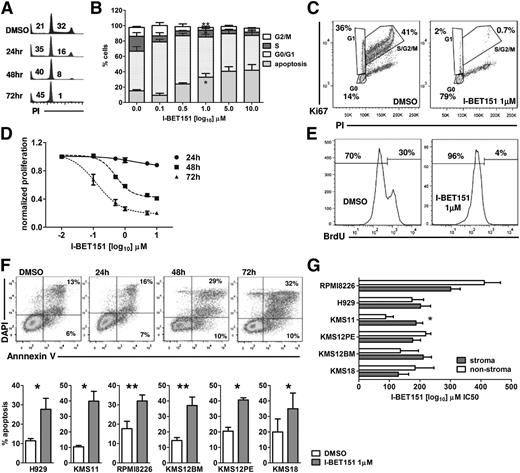
We used DNA content analysis to study the effect of I-BET151 on the cell cycle status of 6 myeloma cell lines (H929, KMS12PE, KMS12BM, KMS18, KMS11, and RPMI8226), representative of the most common translocations found in myeloma (Table 1).19 I-BET151 caused a significant dose- and time-dependent decrease in the proportion of myeloma cells in S/G2 phase at 24, 48, and 72 hours (Figure 1A-B). The most pronounced effect was observed at 72 hours in all 6 myeloma cell lines, starting at 100 nM (supplemental Figure 1A). Dual Ki67/propidium iodide staining confirmed that the majority of live cells resided in the G0 phase after treatment with I-BET151 at 1 μM for 72 hours (Figure 1C; supplemental Figure 1B) commensurate with a dose- and time-dependent decrease in cell proliferation (Figure 1D) and abrogation of bromodeoxyuridine incorporation (Figure 1E).
I-BET151 induces cell cycle arrest and apoptosis in myeloma cell lines. (A) Propidium iodide (PI) staining and flow cytometric cell cycle and apoptosis analysis at indicated time points of the myeloma cell line H929 treated with DMSO or 1 μM I-BET151. (B) Cell cycle analysis of H929 cells after 72-hour culture in the presence of escalating concentrations of I-BET151. Cumulative results of apoptosis induction (subG0/G1) and inhibition of cell proliferation (S/G2 phase). Data are shown as mean ± standard error of the mean (SEM) (n = 3). (C) PI/Ki67 staining and flow cytometric cell cycle analysis of the H929 myeloma cells treated with DMSO or 1 μM I-BET151 for 72 hours. (D) Dose- and time-dependent inhibition of H929 cell proliferation by I-BET151 as assessed by a luminescent proliferation assay. Data are shown as mean ± SEM (n = 3). (E) Flow cytometric assessment of bromodeoxyuridine (BrdU) incorporation by H929 myeloma cells after culture with DMSO or 1 μM I-BET151 for 72 hours. (F) Top: Annexin-V/4',6-diamidino-2-phenylindole (DAPI) staining and flow cytometric analysis of cell viability after treating H929 myeloma cells for the indicated time points with 1 μM I-BE151. Bottom: percent apoptosis (ie, Annexin-V+ cells) in each of the 6 myeloma cell lines at 72 hours after treatment with DMSO or I-BET151. *P < .05, **P < .01. (G) Histograms showing the IC50 of I-BET151 against myeloma cell lines cultured in MS5 cell stroma or stroma-free conditions (mean ± SEM; n = 3), as assessed by a luminescent adenosine triphosphate (ATP) detection-based proliferation assay.
I-BET151 induces cell cycle arrest and apoptosis in myeloma cell lines. (A) Propidium iodide (PI) staining and flow cytometric cell cycle and apoptosis analysis at indicated time points of the myeloma cell line H929 treated with DMSO or 1 μM I-BET151. (B) Cell cycle analysis of H929 cells after 72-hour culture in the presence of escalating concentrations of I-BET151. Cumulative results of apoptosis induction (subG0/G1) and inhibition of cell proliferation (S/G2 phase). Data are shown as mean ± standard error of the mean (SEM) (n = 3). (C) PI/Ki67 staining and flow cytometric cell cycle analysis of the H929 myeloma cells treated with DMSO or 1 μM I-BET151 for 72 hours. (D) Dose- and time-dependent inhibition of H929 cell proliferation by I-BET151 as assessed by a luminescent proliferation assay. Data are shown as mean ± SEM (n = 3). (E) Flow cytometric assessment of bromodeoxyuridine (BrdU) incorporation by H929 myeloma cells after culture with DMSO or 1 μM I-BET151 for 72 hours. (F) Top: Annexin-V/4',6-diamidino-2-phenylindole (DAPI) staining and flow cytometric analysis of cell viability after treating H929 myeloma cells for the indicated time points with 1 μM I-BE151. Bottom: percent apoptosis (ie, Annexin-V+ cells) in each of the 6 myeloma cell lines at 72 hours after treatment with DMSO or I-BET151. *P < .05, **P < .01. (G) Histograms showing the IC50 of I-BET151 against myeloma cell lines cultured in MS5 cell stroma or stroma-free conditions (mean ± SEM; n = 3), as assessed by a luminescent adenosine triphosphate (ATP) detection-based proliferation assay.
Annexin-V staining also confirmed an increased proportion of events in apoptosis (Figure 1F). The delayed rather than immediate effects of I-BET151 on myeloma cell proliferation and survival suggest that these effects are specific and not the result of nonspecific cell toxicity.
We investigated the I-BET151 antimyeloma activity on the MS5 stroma cell line conditions. We found that in 5 of 6 cell lines tested, I-BET151 50% inhibitory concentration (IC50) was similar between stroma and stroma-free culture (P > .05; IC50 range, 133 to 411 nM), while in KMS11, IC50 increased from 88 to 186 nM in the presence of stroma (P = .002; Figure 1G).
Activity of I-BET151 against primary myeloma cells in vitro and in xenotransplantation
Next, we determined the antimyeloma activity of I-BET151 on CD138-selected primary BM myeloma cells (n = 4 patients) cultured on MS5 stroma in the presence of IL-6 (5 ng/mL). As in myeloma cell lines, there was a dose-dependent decrease in the proportion of cells in S/G2 phase and an increase of subG0 events, suggesting that myeloma cells were entering cell cycle arrest and undergoing apoptosis (Figure 2A-B). In addition, in an in vivo model of subcutaneous myeloma, I-BET151–treated mice had four- to fivefold smaller myeloma tumors (P < .001; Figure 2C) and a significantly reduced rate of tumor size doubling than vehicle-treated mice (P < .001; Figure 2D). We concluded that I-BET151 has significant antimyeloma activity in vitro and in vivo, even in the presence of stroma and promyeloma survival factors.
Antimyeloma activity of I -BET151 against stroma and growth factor–treated myeloma cell lines and primary myeloma cells in vitro and in vivo. (A-B) Representative examples and cumulative data of (A) cell cycle and (B) apoptosis analysis of primary CD138+ myeloma cells cultured on MS5 stroma cells and in the presence of IL-6 5 ng/mL and treated with DMSO or I-BET151 for 72 hours (n = 4). Data shown as mean ± SEM. (C) KMS11 cells (5 × 106) were injected subcutaneously into the flank of NSG mice. Treatment with I-BET151 (n = 13; 30 mg/kg intraperitoneal injection daily for 21 consecutive days) or vehicle (n = 11; 0.9% NaCl + 10% kleptose + 5% DMSO) was initiated when the maximum dimension of the tumor was ≥5 mm. Relative increase of tumor volume compared with day 1 of treatment is shown. (D) Kaplan-Meier survival analysis of time to progression, defined as time to doubling of tumor size compared with day 1 of treatment. Log-rank test P < .001.
Antimyeloma activity of I -BET151 against stroma and growth factor–treated myeloma cell lines and primary myeloma cells in vitro and in vivo. (A-B) Representative examples and cumulative data of (A) cell cycle and (B) apoptosis analysis of primary CD138+ myeloma cells cultured on MS5 stroma cells and in the presence of IL-6 5 ng/mL and treated with DMSO or I-BET151 for 72 hours (n = 4). Data shown as mean ± SEM. (C) KMS11 cells (5 × 106) were injected subcutaneously into the flank of NSG mice. Treatment with I-BET151 (n = 13; 30 mg/kg intraperitoneal injection daily for 21 consecutive days) or vehicle (n = 11; 0.9% NaCl + 10% kleptose + 5% DMSO) was initiated when the maximum dimension of the tumor was ≥5 mm. Relative increase of tumor volume compared with day 1 of treatment is shown. (D) Kaplan-Meier survival analysis of time to progression, defined as time to doubling of tumor size compared with day 1 of treatment. Log-rank test P < .001.
MYC-dependent and -independent molecular signatures of I-BET151 antimyeloma activity
Much of the antiproliferative effect of the BET domain inhibitors has been attributed to direct inhibition of transcription of MYC and consequently of the MYC-dependent transcriptional program.11,14,15 To investigate the molecular basis of I-BET151 antimyeloma activity, we generated a global RNA profile of the myeloma cell lines H929 and KMS12BM which carry t(4,14) and t(11;14), respectively, two of the most common primary genetic events in myeloma.2,3 Because the transcriptional programs activated by the two oncogenic translocations differ23 and in order to study the coordinated changes in biologic networks associated with each phenotype, we performed GSEA of the GEP data from each cell line individually.
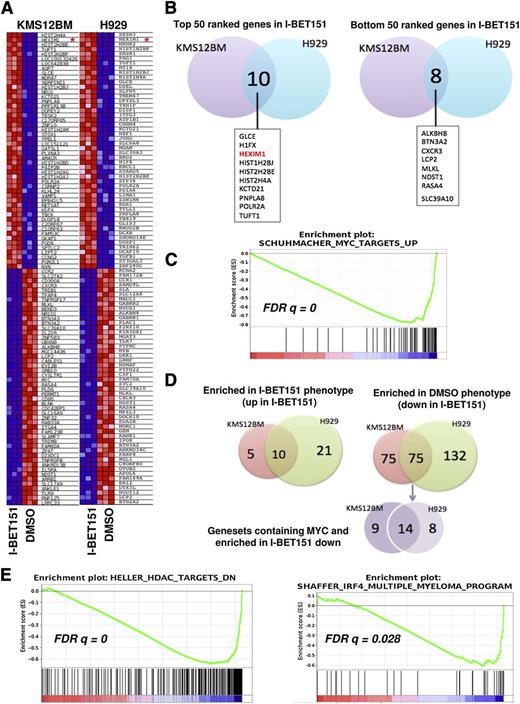
We found that after treatment with I-BET151, among the top (most upregulated) and bottom (most downregulated) 50 genes in the gene ranking metrics (Figure 3A), only around 10% are overlapping between the two cell lines (Figure 3B), probably reflecting their distinct oncogenic programs and genetic profiles.
MYC-dependent and -independent molecular signatures of I-BET151 antimyeloma activity. (A) Heat map showing the 50 most up- and downregulated genes after treatment with I-BET151 (1 μM) for 6 hours of KMS12BM and H929 cells. HEXIM1 is highlighted by a red asterisk. (B) Venn diagram showing that among the 50 most upregulated genes in I-BET151-treated cells, 10 are shared by the 2 cell lines (left), while among the 50 most downregulated genes, 8 are shared by the 2 cell lines (right). (C) A representative MYC-dependent gene set enriched in both cell lines as determined by GSEA. (D) Venn diagrams showing the number of gene sets significantly enriched (P < .01; FDR q < 0.05) either in both or in individual cell lines. Left: number of gene sets significantly enriched among list of genes upregulated in I-BET151-treated cells; right: number of gene sets enriched among list of genes downregulated in I-BET151-treated cells. MYC-dependent gene sets enriched in the most downregulated genes in response to I-BET151 (shown in purple) form only a minority, with the majority of enriched gene sets being MYC-independent (supplemental Table 1). (E) GSEA analysis reveals enrichment of histone deacetylase (HDAC) inhibitor signatures in both H929 and KMS12BM cell lines (left; only KMS12BM shown here), and a myeloma-specific IRF4-dependent gene set in KMS12BM but not H929 cells (right) upon treatment with I-BET151.
MYC-dependent and -independent molecular signatures of I-BET151 antimyeloma activity. (A) Heat map showing the 50 most up- and downregulated genes after treatment with I-BET151 (1 μM) for 6 hours of KMS12BM and H929 cells. HEXIM1 is highlighted by a red asterisk. (B) Venn diagram showing that among the 50 most upregulated genes in I-BET151-treated cells, 10 are shared by the 2 cell lines (left), while among the 50 most downregulated genes, 8 are shared by the 2 cell lines (right). (C) A representative MYC-dependent gene set enriched in both cell lines as determined by GSEA. (D) Venn diagrams showing the number of gene sets significantly enriched (P < .01; FDR q < 0.05) either in both or in individual cell lines. Left: number of gene sets significantly enriched among list of genes upregulated in I-BET151-treated cells; right: number of gene sets enriched among list of genes downregulated in I-BET151-treated cells. MYC-dependent gene sets enriched in the most downregulated genes in response to I-BET151 (shown in purple) form only a minority, with the majority of enriched gene sets being MYC-independent (supplemental Table 1). (E) GSEA analysis reveals enrichment of histone deacetylase (HDAC) inhibitor signatures in both H929 and KMS12BM cell lines (left; only KMS12BM shown here), and a myeloma-specific IRF4-dependent gene set in KMS12BM but not H929 cells (right) upon treatment with I-BET151.
In agreement with previous data,11,14,15 MYC was among the 20 most downregulated genes in KMS12BM cells (Figure 3A-B) and among the 7% most downregulated genes in H929 cells (not shown). In both cell lines, MYC downregulation was confirmed by RQ-PCR (supplemental Figure 2A). Upon GSEA, 4 MYC-dependent gene sets were significantly (FDR q < .05) enriched in association with downregulated expression by I-BET151 in both cell lines24 (Figure 3C; supplemental Table 1). Genes with MYC and MAX (a transcriptional partner of MYC) DNA binding motifs were also significantly downregulated in KMS12BM but not H929 cells (supplemental Figure 2B).
Since not all tumor cells with MYC overexpression are sensitive to BET inhibitors and since MYC expression is not always downregulated by BET domain inhibition,10,11 alternative MYC-independent mechanisms could also underpin the observed antiproliferative effect. We identified a number of MYC-dependent but also a much larger number of MYC-independent molecular signatures that were enriched in both or in one of the two cell lines (Figure 3D; supplemental Table 1). These included a gene set representing genes previously downregulated in myeloma cell lines treated with the histone deacetylase inhibitor Trichostatin25 (Figure 3E), a finding previously reported in GEP studies involving JQ1.15 IRF4-dependent signature was downregulated in KMS12BM (Figure 3E; supplemental Table 1). IRF4 is a transcription factor critical for normal plasma cell development,26 but it also drives a myeloma-specific transcriptional program indispensable for cell survival.27
I-BET151 inhibits transcription of oncogenic MYC by interfering with its BRD4–dependent transcriptional activation
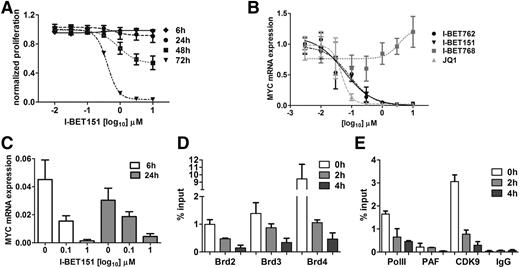
To explore the transcriptional events in MYC downregulation, we used OPM-2 myeloma cells characterized by MYC overexpression that results from a translocation that brings MYC under the transcriptional control of immunoglobulin H (IGH) gene enhancers.28,29 As for the other myeloma cell lines, I-BET151 caused a time- and dose-dependent inhibition of OPM-2 cell proliferation (Figure 4A) with commensurate reduction in MYC messenger RNA (mRNA) expression (Figure 4B-C). Next we studied the cis and trans changes at the IGH enhancer that drives MYC overexpression in OPM-2 cells. We specifically focused on the role of BRD2-4 (expression of BRDT is restricted to testis and ovary), RNA Pol II, P-TEFb, and PAF complexes. We found that upon treatment with I-BET151, BRD2, -3, and -4 occupancy decreased in a time-dependent manner as early as 2 hours posttreatment (Figure 4D). Similarly, recruitment of CDK9 and PAF, critical components of the P-TEFb and PAFc1 complexes, respectively, as well as binding of RNA Pol II to the IGH gene enhancer, were almost abolished (Figure 4E). We conclude that I-BET151–mediated transcriptional silencing of MYC in myeloma cells involves inhibition of BRD2-4 binding, failure to recruit P-TEFb and PAFc1 complexes to the IGH enhancer by BRD2-4, and lack of RNA Pol II recruitment.
I-BET151 inhibits transcription of oncogenic MYC by interfering with BRD4-dependent transcriptional activation. (A) Time- and dose-dependent effect of I-BET151 on OPM-2 myeloma cell proliferation (n = 3). (B) MYC transcript levels after 6 hours of treatment with different doses of I-BET151 and I-BET762, their inactive enantiomer I-BET768, and JQ1. Data were normalized to vehicle control (n = 3). (C) MYC transcript levels after 6 or 24 hours of treatment with two different doses of I-BET151 or vehicle (n = 3). (D) chromatin immunoprecipitation (ChIP)–RQ-PCR analysis of BRD2, -3 and -4 binding onto the IGH enhancer in OPM-2 cells at 0, 2, and 4 hours after treatment with 1 μM I-BET151. Data are shown as percent of input (n = 3). (E) ChIP–RQ-PCR analysis of CDK9, PAF, and RNA Pol II binding onto the IGH enhancer of OPM-2 cells. Background IgG binding as percentage of input for ChIP assays in (D) and (E) is also shown (n = 3). Data are shown as mean ± SEM.
I-BET151 inhibits transcription of oncogenic MYC by interfering with BRD4-dependent transcriptional activation. (A) Time- and dose-dependent effect of I-BET151 on OPM-2 myeloma cell proliferation (n = 3). (B) MYC transcript levels after 6 hours of treatment with different doses of I-BET151 and I-BET762, their inactive enantiomer I-BET768, and JQ1. Data were normalized to vehicle control (n = 3). (C) MYC transcript levels after 6 or 24 hours of treatment with two different doses of I-BET151 or vehicle (n = 3). (D) chromatin immunoprecipitation (ChIP)–RQ-PCR analysis of BRD2, -3 and -4 binding onto the IGH enhancer in OPM-2 cells at 0, 2, and 4 hours after treatment with 1 μM I-BET151. Data are shown as percent of input (n = 3). (E) ChIP–RQ-PCR analysis of CDK9, PAF, and RNA Pol II binding onto the IGH enhancer of OPM-2 cells. Background IgG binding as percentage of input for ChIP assays in (D) and (E) is also shown (n = 3). Data are shown as mean ± SEM.
Dynamics of I-BET151–induced transcription of HEXIM1
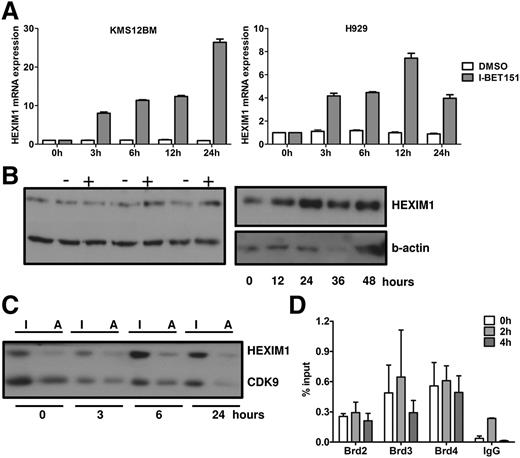
Our GEP analysis showed that HEXIM1, a negative regulator of P-TEFb,30-32 was one of the most upregulated genes in both cell lines after treatment with I-BET151 (Figure 3A), as confirmed with RQ-PCR (Figure 5A). Interestingly, HEXIM1 is also within the top 10 most upregulated genes in lymphoma and myeloma cell lines (different from those reported here) treated with JQ1,14,15 suggesting that HEXIM1 upregulation is a consistent feature of BET protein inhibitors. While HEXIM1 mRNA levels increased as early as 3 hours after I-BET151 treatment, increased HEXIM1 protein levels were first detected in H929 cells at 6 hours posttreatment (Figure 5B-C). A similar pattern was seen in OPM-2 and KMS12BM cells (supplemental Figure 3A).
Role of HEXIM1 in the antiproliferative effect of I-BET151. (A) RQ-PCR analysis of MYC and HEXIM1 mRNA expression in H929 and KMS12BM myeloma cells over 24 hours after treatment with I-BET151 (1 μM) or DMSO control. Expression is normalized against the RNA levels at t = 0. Data shown are representative of 2 independent experiments; mean ± SEM of technical triplicate assays for each time point is shown. (B) HEXIM1 protein levels in H929 myeloma cells as assessed by immunoblotting after treatment with I-BET151 1 μM (+) or DMSO (−) control at the time points indicated. Two immunoblots with different time points are shown. (C) Relative levels of active (HEXIM1-free) and inactive (HEXIM1-bound) CDK9 upon treatment with I-BET151 (1 μM) of H929 cells. Following differential salt extraction of nuclei and running of low- (inactive CDK9 fraction) and high-salt (active CDK9 fraction) extracts for each time point side-by-side on sodium dodecyl sulfate polyacrylamide gel electrophoresis, the same membrane was immunoblotted with anti-HEXIM1 and CDK9 antibody. (D). ChIP assay performed under the same conditions described in Figure 4, showing occupancy at HEXIM1 promoter of BRD2-4 at 0, 2, and 4 hours after treatment with I-BET151 (1 μM; n = 3). I, inactive CDK9; A, active CDK9.
Role of HEXIM1 in the antiproliferative effect of I-BET151. (A) RQ-PCR analysis of MYC and HEXIM1 mRNA expression in H929 and KMS12BM myeloma cells over 24 hours after treatment with I-BET151 (1 μM) or DMSO control. Expression is normalized against the RNA levels at t = 0. Data shown are representative of 2 independent experiments; mean ± SEM of technical triplicate assays for each time point is shown. (B) HEXIM1 protein levels in H929 myeloma cells as assessed by immunoblotting after treatment with I-BET151 1 μM (+) or DMSO (−) control at the time points indicated. Two immunoblots with different time points are shown. (C) Relative levels of active (HEXIM1-free) and inactive (HEXIM1-bound) CDK9 upon treatment with I-BET151 (1 μM) of H929 cells. Following differential salt extraction of nuclei and running of low- (inactive CDK9 fraction) and high-salt (active CDK9 fraction) extracts for each time point side-by-side on sodium dodecyl sulfate polyacrylamide gel electrophoresis, the same membrane was immunoblotted with anti-HEXIM1 and CDK9 antibody. (D). ChIP assay performed under the same conditions described in Figure 4, showing occupancy at HEXIM1 promoter of BRD2-4 at 0, 2, and 4 hours after treatment with I-BET151 (1 μM; n = 3). I, inactive CDK9; A, active CDK9.
Since the main function of HEXIM1 is to sequester and inactivate P-TEFb (ie, CDK9 and cyclin T1), we analyzed the relative amount of inactive and active P-TEFb fractions. By using a previously validated methodology33,34 (supplemental Figure 3), we assessed the relative amounts of HEXIM1-bound inactive and HEXIM1-free active CDK9 in the low- and high-salt cell extracts, respectively, by immunoblotting after treatment with I-BET151 (Figure 5C; supplemental Figure 3B).
Overall, we found that in H929 cells, after a temporary increase, active CDK9 eventually decreased to levels lower than baseline (Figure 5C) in the first 24 hours following treatment with I-BET151. By contrast, a decrease in the levels of active CDK9 in OPM-2 cells was observed only beyond 48 hours when apoptosis and cell cycle arrest were first observed (supplemental Figure 3). These findings suggest that, at least in the case of H929 cells, increased HEXIM1 expression in response to I-BET treatment is associated with reduced availability of active CDK9 and, consequently, reduced availability of P-TEFb to participate in active transcription.
To investigate the role of BRD2-4 in HEXIM1 transcription in response to I-BET151, we performed chromatin immunoprecipitation analysis of BRD2-4 binding at the promoter of HEXIM1 at the same time in the same OPM-2 cells in which I-BET151 induced abrogation of BRD2-4 binding at the IGH enhancer (Figure 3). We found enrichment of BRD2-4 in untreated cells but at lower levels than at the IGH enhancer (Figure 3). However, in contrast to the IGH enhancer, BRD2-4 occupancy did not change in response to I-BET151, suggesting that BRD2-4 binding at the promoter of HEXIM1 is insensitive to I-BET151 (Figure 5D). However, cyclohexamide treatment had no significant effect on I-BET–mediated increase in HEXIM1 mRNA levels, suggesting that HEXIM1 mRNA upregulation is a direct consequence of I-BET151 treatment (supplemental Figure 3C).
Taken together, these data show that HEXIM1 overexpression in response to I-BET151 is independent of BRD2-4 but leads to reduced availability of the active form of P-TEFb that is responsible for transcriptional initiation and elongation.
In vitro and in vivo antimyeloma activity of oral I-BET762
The preclinical tool compound I-BET151 has previously been shown to have antileukemia activity,11 and our data show that it is also active against myeloma. On the basis of these observations, we then tested the antimyeloma activity of I-BET762, a compound with a pharmacologic profile similar to that of I-BET151 but with suitable properties for clinical development.20 As shown in Figure 6A, compared with the inactive enantiomer I-BET768, I-BET762 inhibited OPM-2 cell proliferation with the same kinetics and comparable IC50 as I-BET151 and JQ1 and also inhibited MYC transcription (Figure 4B). I-BET762 exerted an antiproliferative effect on several other myeloma cell lines, as well as on OPM-2, with a growth IC50 <1.0 µM observed in the majority of these lines (71%; 10 of 14) (Table 1).
In vitro and in vivo antimyeloma activity of I-BET 762. (A) Day 1 and 2 OPM-2 myeloma cell line proliferation in response to different concentrations of I-BET762, I-BET151, and JQ1 as assessed in 3H-thymidine incorporation assays. I-BET762 is also compared against the inactive I-BET768 compound. Y-axis depicts normalized proliferation (n = 3). Data are shown as mean ± SEM. (B) Body weight of mice treated with different dosing schemes of I-BET762 or vehicle control. Day 1 corresponds to day 15 (ie, when I-BET762 administration commenced). Data are expressed as mean ± SEM (n = 15) for the start of each dosing schedule. (C) Peripheral blood hLC concentration at time points shown in mice treated with 4 different dosing schemes of I-BET762 (3 mg/kg per day, 10 mg/kg per day, 30 mg/kg once every other day [ie, alternate days], and 30 to 20 mg/kg per day [ie, 30 mg/kg for 15 days, followed by 2 weeks off treatment, followed by 20 mg/kg until termination of the experiment starting from day 15 after myeloma cell inoculation] or vehicle control (n = 8 to 11 mice per dosing schedule). Samples from one animal treated with vehicle and one animal treated with 20 to 30 mg/kg were taken at days 54 and 62, respectively, but presented in the day 58 group. Data are shown as mean ± SEM. *P < .05; **P < .01; ***P < .001. (D) Frequency of hCD38+ OPM-2 human myeloma cells in the BM taken from the femur as assessed by flow cytometry at the time of euthanasia. Data are shown as mean ± SEM (n = 11 to 15). (E) Kaplan-Meier survival analysis of OPM-2–bearing mice in different treatment groups (n = 8 to 11 mice per dosing schedule) for each of the I-BET762 treatment groups against vehicle-treated group; log-rank test P ≤ .002. nd, not done.
In vitro and in vivo antimyeloma activity of I-BET 762. (A) Day 1 and 2 OPM-2 myeloma cell line proliferation in response to different concentrations of I-BET762, I-BET151, and JQ1 as assessed in 3H-thymidine incorporation assays. I-BET762 is also compared against the inactive I-BET768 compound. Y-axis depicts normalized proliferation (n = 3). Data are shown as mean ± SEM. (B) Body weight of mice treated with different dosing schemes of I-BET762 or vehicle control. Day 1 corresponds to day 15 (ie, when I-BET762 administration commenced). Data are expressed as mean ± SEM (n = 15) for the start of each dosing schedule. (C) Peripheral blood hLC concentration at time points shown in mice treated with 4 different dosing schemes of I-BET762 (3 mg/kg per day, 10 mg/kg per day, 30 mg/kg once every other day [ie, alternate days], and 30 to 20 mg/kg per day [ie, 30 mg/kg for 15 days, followed by 2 weeks off treatment, followed by 20 mg/kg until termination of the experiment starting from day 15 after myeloma cell inoculation] or vehicle control (n = 8 to 11 mice per dosing schedule). Samples from one animal treated with vehicle and one animal treated with 20 to 30 mg/kg were taken at days 54 and 62, respectively, but presented in the day 58 group. Data are shown as mean ± SEM. *P < .05; **P < .01; ***P < .001. (D) Frequency of hCD38+ OPM-2 human myeloma cells in the BM taken from the femur as assessed by flow cytometry at the time of euthanasia. Data are shown as mean ± SEM (n = 11 to 15). (E) Kaplan-Meier survival analysis of OPM-2–bearing mice in different treatment groups (n = 8 to 11 mice per dosing schedule) for each of the I-BET762 treatment groups against vehicle-treated group; log-rank test P ≤ .002. nd, not done.
Next, we tested the antimyeloma activity of I-BET762 dosed orally in an in vivo systemic xenograft model generated by injecting OPM-2 cells into NOD-SCID mice. Daily oral doses of I-BET762 up to 10 mg/kg and 30 mg/kg given every other day were well tolerated with no clear impact on body weight compared with vehicle control (Figure 6B). We found that plasma hLC concentration was significantly reduced in mice treated with I-BET762 (Figure 6C). Specifically, as disease progressed, hLC concentration in the blood of myeloma-bearing mice increased precipitously. As expected, in vehicle-treated animals, levels of hLC continued to increase until termination, consistent with progressive myeloma. Although an increase in hLC levels was found in mice treated with I-BET762, mice treated with the 3 highest doses showed a significant reduction (P ≤ .001) in the hLC concentration at all 4 time points studied (Figure 6C). Human CD38+ BM cells were 10% in vehicle-treated animals, while they were <1% in animals treated with the 3 highest doses (P ≤ .001) (Figure 6D; supplemental Figure 4A). Similarly, histopathologic analysis of vertebrae at the time of euthanasia shows significantly lower OPM-2 cell infiltration in I-BET762–treated animals (supplemental Figure 4B). Finally, pharmacokinetic sampling 30 minutes after dose in this study was consistent with anticipated concentrations based on studies of intravenous or oral administration at 3 and 30 mg/kg in BALB/c mice (supplemental Methods and supplemental Table 2).
This considerable antimyeloma activity resulted in a significant (P ≤ .002) survival advantage observed in all 4 I-BET762–treated groups of mice, with median survival not reached in animals treated with the 3 highest doses of I-BET762 (Figure 6E), notably including the groups of mice dosed at 20 to 30 mg/kg per day (that had a dosing holiday during the study) and those at 30 mg/kg every other day (Figure 6E). These data represent the first example of an orally active BET inhibitor significantly delaying myeloma progression in vivo.
Discussion
This study provides mechanistic insight and extended preclinical evidence that I-BET151 and I-BET762, representing the quinoline and benzodiazepine classes of BET inhibitors, respectively, possess considerable antimyeloma activity in vitro and in vivo. I-BET151 and I-BET762 constitute novel small molecules that interfere with the ability of the BET family of epigenetic “readers” to modulate transcription.12 JQ1 was previously shown to exert antimyeloma activity through cell cycle inhibition and senescence but not apoptosis.14 We found that, similar to JQ1, both I-BET151 and I-BET762 also induce cell cycle arrest and thus exert a potent antiproliferative effect against myeloma. In addition, they have a clear proapoptotic effect, as previously described, against acute myeloid leukemia cells that is linked to downregulation of the prosurvival factor BCL2,11 also observed in both myeloma cell lines treated with I-BET151 (data not shown).
Downregulation of MYC transcription and of the MYC oncogenic programs have been shown to be important pathways mediating the antiproliferative effect of BET inhibitors in many hematologic malignancies, including in animal models of myeloma treated with JQ1.35 In GEP data analysis previously reported for JQ1,14,15 all cell lines were considered together in two homogeneous groups (ie, before and after treatment with JQ1). To capture the overlapping but also distinct oncogenic programs that reflect the distinct myeloma-initiating genetic events,23 we analyzed our GEP data separately for each cell line. Through this approach, we found that I-BET151, similar to JQ1, caused MYC and MYC-dependent gene transcription downregulation in both myeloma cell lines. At a mechanistic level, by using the MYC/IGH rearrangement in OPM-2 cells as an example of oncogenic MYC activation, we show that in response to I-BET151, MYC transcription is abrogated by inhibition of BRD2-4 binding to the IGH enhancer, leading to decreased CDK9, PAF, and RNA Pol II recruitment.
A consistent feature of all BET inhibitors is their ability to induce overexpression of HEXIM1, a negative regulator of pTEFb,30-32 in different hematologic tumors (including myeloma).14,15 HEXIM1 is part of a ribonucleoprotein complex that consists of cyclin T1/2, CDK9, and small nuclear RNA 7SK, which together form the catalytically inactive form of pTEFb.30-32,36 By contrast, catalytically active P-TEFb requires dissociation from HEXIM1 and its recruitment via BRD4 or transcription factors to the target genes where, through CDK9-dependent Ser2 phosphorylation of the carboxyterminal domain of RNA Pol II, it promotes transcriptional activation and elongation.30-32,36
Our biochemical dissection of the equilibrium of active versus inactive P-TEFb suggests that, at least in H929 cells, availability of active CDK9 eventually decreases in response to I-BET151 in the first 24 hours. Interestingly, the same pattern associated with HEXIM1 overexpression was recently reported for JQ1,34 suggesting that BET inhibitors share the same mechanism of HEXIM1 regulation. However, transcriptional activation of HEXIM1 in response to I-BET151 is not associated with changes in BRD2-4 binding and therefore HEXIM1 transcription is BET protein–independent, suggesting that the ability of BET inhibitors to prevent BRD2-4 binding to acetylated histones is not uniform.
The limited availability of active P-TEFb as a result of sustained overexpression of HEXIM1 in myeloma cells in response to I-BET151 would be expected to have a significant impact on the transcription of a large number of genes, including oncogenic MYC, that require transcription factor- or BRD4-mediated recruitment of P-TEFb for their transcriptional activation.9 Interestingly, MYC itself leads to Pol II activation through recruitment of P-TEFb to chromatin37 ; therefore, I-BET151–mediated HEXIM1 overexpression could contribute to silencing of genes that are regulated by MYC-mediated recruitment of P-TEFb, a notion that will require further experimental exploration. Because HEXIM1 knockdown resulted in prompt myeloma cell death (data not shown), we were unable to directly test how HEXIM1 might cooperate at the genetic level with I-BET151 in affecting cell myeloma proliferation and survival.
Importantly, we assessed the IC50 of I-BET151 on myeloma cells grown on stroma, thereby simulating the in vivo dependence of myeloma cells on and their interaction with their microenvironment.38 Since myeloma cell survival, proliferation, and drug resistance have all been shown to be enhanced by microenvironmental cues,39 it is significant that the I-BET151 IC50 was not affected by the presence of stroma in most cell lines. The potent antimyeloma activity of I-BET151 was also apparent in stroma-dependent cultures of primary myeloma cells, even in the presence of IL-6, a growth factor that is essential for myeloma survival in vivo.40-42 Pertinent to this, IL-6 receptor mRNA levels were significantly decreased in both myeloma cell lines in response to I-BET151 (data not shown).
Finally, we demonstrate that a significant survival advantage is gained by I-BET762 in the treatment of established, systemic myeloma in mice, consistent with its in vitro antimyeloma activity. These data provide the first evidence that I-BET762,13 currently in phase 1 studies in solid tumors,43 has considerable preclinical antimyeloma activity. Importantly, this drug-like small molecule with good oral pharmacokinetics (supplemental Table 2) highlights for the first time the promise of a BET inhibitor as an effective oral antimyeloma agent. Previous reports with the chemical probe molecule JQ1 have relied on parenteral dosing to obtain adequate drug exposures to drive efficacy and indeed, to date, there are no reports of this molecule progressing further into clinical development.
In conclusion, we showed that the tool compound I-BET151 and the structurally distinct clinical candidate I-BET762 demonstrate significant preclinical antimyeloma activity associated with contrasting effects on MYC and HEXIM1 transcription. Our data provide clear rationale for extending the clinical evaluation of the novel therapeutic agent I-BET762 in multiple myeloma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
OPM-2 systemic myeloma xenograft was performed by contract research provider CrownBio, Beijing, China.
This work was supported by Leukaemia and Lymphoma Research, Leuka, and the National Institute for Health Research Biomedical Research Centre.
Authorship
Contribution: A.C. performed research, analyzed data, and wrote the manuscript; B. Liu analyzed gene expression profiling data; I.M., M.S.C., V.C., K.G., A. Rotolo, D.F.T., A.K.B., T.D.C., N.R.H., O.B., P.T., N.A.-M., A.C.H., L.C., and B. Le performed research and analyzed data; J.J.W., N.N.S., N.J.P., and R.K.P. designed and supervised research and contributed to writing the manuscript; A. Rahemtulla commented on the manuscript and provided clinical samples; I.R. and M.K. supervised research and commented on the manuscript; and A.K. supervised research, analyzed data, and wrote the first draft of the manuscript.
Conflict-of-interest disclosure: D.F.T., N.N.S., A.K.B., T.D.C., N.R.H., O.B., P.T., N.A.-M., A.C.H., L.C., B. Le, J.J.W., N.J.P., and R.K.P. are employees of GlaxoSmithKline. The remaining authors declare no competing financial interests.
Correspondence: Anastasios Karadimitris, Centre for Haematology, Imperial College London, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: a.karadimitris@imperial.ac.uk.



![Figure 6. In vitro and in vivo antimyeloma activity of I-BET 762. (A) Day 1 and 2 OPM-2 myeloma cell line proliferation in response to different concentrations of I-BET762, I-BET151, and JQ1 as assessed in 3H-thymidine incorporation assays. I-BET762 is also compared against the inactive I-BET768 compound. Y-axis depicts normalized proliferation (n = 3). Data are shown as mean ± SEM. (B) Body weight of mice treated with different dosing schemes of I-BET762 or vehicle control. Day 1 corresponds to day 15 (ie, when I-BET762 administration commenced). Data are expressed as mean ± SEM (n = 15) for the start of each dosing schedule. (C) Peripheral blood hLC concentration at time points shown in mice treated with 4 different dosing schemes of I-BET762 (3 mg/kg per day, 10 mg/kg per day, 30 mg/kg once every other day [ie, alternate days], and 30 to 20 mg/kg per day [ie, 30 mg/kg for 15 days, followed by 2 weeks off treatment, followed by 20 mg/kg until termination of the experiment starting from day 15 after myeloma cell inoculation] or vehicle control (n = 8 to 11 mice per dosing schedule). Samples from one animal treated with vehicle and one animal treated with 20 to 30 mg/kg were taken at days 54 and 62, respectively, but presented in the day 58 group. Data are shown as mean ± SEM. *P < .05; **P < .01; ***P < .001. (D) Frequency of hCD38+ OPM-2 human myeloma cells in the BM taken from the femur as assessed by flow cytometry at the time of euthanasia. Data are shown as mean ± SEM (n = 11 to 15). (E) Kaplan-Meier survival analysis of OPM-2–bearing mice in different treatment groups (n = 8 to 11 mice per dosing schedule) for each of the I-BET762 treatment groups against vehicle-treated group; log-rank test P ≤ .002. nd, not done.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/5/10.1182_blood-2013-01-478420/4/m_697f6.jpeg?Expires=1767812162&Signature=IR4x6hXZMO270ZVdKlA7F~qZs8yejnJ7aCvztbCN9laROZIhsFypEcCHGhGxtaQ5eVakArccFgeBzkWO3s3LDPBl5EVbGcK537QNmIt4bkFkTjGEaZSPaxTTCZzttnWzAClmXvOOrRAtRzxOOCIJtcMS7VZNu0YIdrWx~jK1UuummjGR4yH6KuZP3M-G0UkGuW9kdRApcwnAxhmT9hOZT1vAaPgjdVgBb20hYNVnjWHw-FRSBIlHGQHDiJr~-FlgI9fdM9JXMDps2iVKlgBFV6zoD7zajulabRPVa-aBGNe4qFznh8nDgu8Ktt2z41Kg5uoSvM8jFMF2EH1t1VPzYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 6. In vitro and in vivo antimyeloma activity of I-BET 762. (A) Day 1 and 2 OPM-2 myeloma cell line proliferation in response to different concentrations of I-BET762, I-BET151, and JQ1 as assessed in 3H-thymidine incorporation assays. I-BET762 is also compared against the inactive I-BET768 compound. Y-axis depicts normalized proliferation (n = 3). Data are shown as mean ± SEM. (B) Body weight of mice treated with different dosing schemes of I-BET762 or vehicle control. Day 1 corresponds to day 15 (ie, when I-BET762 administration commenced). Data are expressed as mean ± SEM (n = 15) for the start of each dosing schedule. (C) Peripheral blood hLC concentration at time points shown in mice treated with 4 different dosing schemes of I-BET762 (3 mg/kg per day, 10 mg/kg per day, 30 mg/kg once every other day [ie, alternate days], and 30 to 20 mg/kg per day [ie, 30 mg/kg for 15 days, followed by 2 weeks off treatment, followed by 20 mg/kg until termination of the experiment starting from day 15 after myeloma cell inoculation] or vehicle control (n = 8 to 11 mice per dosing schedule). Samples from one animal treated with vehicle and one animal treated with 20 to 30 mg/kg were taken at days 54 and 62, respectively, but presented in the day 58 group. Data are shown as mean ± SEM. *P < .05; **P < .01; ***P < .001. (D) Frequency of hCD38+ OPM-2 human myeloma cells in the BM taken from the femur as assessed by flow cytometry at the time of euthanasia. Data are shown as mean ± SEM (n = 11 to 15). (E) Kaplan-Meier survival analysis of OPM-2–bearing mice in different treatment groups (n = 8 to 11 mice per dosing schedule) for each of the I-BET762 treatment groups against vehicle-treated group; log-rank test P ≤ .002. nd, not done.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/5/10.1182_blood-2013-01-478420/4/m_697f6.jpeg?Expires=1767923190&Signature=rALNttRucM39kO~~bxksStKh-GwvMoELx9DGBlQlYK3f9gzQ5w2SZi9829iUY6usQkIrf-izmKQF1uGH8HwSz7DEU75aHNPUhhh~JSloyvO1rkW9AP77Xt47EU8eD-1ien1~Jz-DW1ds9U0fAak3~z116hQtnOiDs~v7ZWN0Q450AkddNo7ym2f34n1dUu7RQfsW9UOjypgBz9s7L8H6-L9KjI8NXcNQUvJ3OXKJeHUJao5gE9bvo9H9g92h5Ny88lwrCBqL15Hi74EwFD40YYJhW5NPhqxxAIjE3sPOs5ZMLcdunkjCxtEL~Si2~QcsBrvC3i-x0BmKuKabpM2esg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)