Key Points
miRNA-130a is expressed in myeloblasts and promyelocytes and inhibits translation of CEBPE mRNA encoding transcription factor C/EBP-ε.
Regulation of CEBPE mRNA by miRNA-130a is required for timed expression of secondary granule proteins and cell cycle exit.
CCAAT/enhancer binding protein-ε (C/EBP-ε) is considered a master transcription factor regulating terminal neutrophil maturation. It is essential for expression of secondary granule proteins, but it also regulates proliferation, cell cycle, and maturation during granulopoiesis. Cebpe-/- mice have incomplete granulocytic differentiation and increased sensitivity toward bacterial infections. The amount of C/EBP-ε messenger RNA (mRNA) increases with maturation from myeloblasts with peak level in myelocytes (MC)/metamyelocytes (MM), when the cells stop proliferating followed by a decline in more mature cells. In contrast, C/EBP-ε protein is virtually detectable only in the MC/MM population, indicating that expression in more immature cells could be inhibited by microRNAs (miRNAs). We found that miRNA-130a (miR-130a) regulates C/EBP-ε protein expression in both murine and human granulocytic precursors. Overexpression of miR-130a in a murine cell line downregulated C/EBP-ε protein and lactoferrin (Ltf), cathelicidin antimicrobial protein (Camp), and lipocalin-2 (Lcn2) mRNA expression giving rise to cells with a more immature phenotype, as seen in the Cebpe−/− mouse. Introduction of a C/EBP-ε mRNA without target site for miR-130a restored both C/EBP-ε production, expression of Camp and Lcn2, and resulted in the cells having a more mature phenotype. We conclude that miR-130a is important for the regulation of the timed expression of C/EBP-ε during granulopoiesis.
Introduction
Granulopoiesis, the development of mature neutrophil granulocytes from myeloblasts (MBs), is a strictly regulated process requiring action of several transcription factors to control processes, such as cell division, differentiation, and correct timing of synthesis of the different granule proteins.1 Dysregulation of transcription factors can be leukemogenic, leading to differentiation arrest and a malignant phenotype, such as acute myeloid leukemia (AML),2 or can result in neutrophils that are not equipped with their full armory for combating invading microorganisms, as seen in specific granule deficiency (SGD), and recapitulated in the phenotype of Cebpe−/− mice.3
CCAAT/enhancer binding protein-ε (C/EBP-ε) is a transcription factor essential for terminal granulopoiesis.4 The C/EBPs constitute a family of highly conserved transcription factors with 6 members (C/EBP-α, -β, -δ, -ε, -γ, and -ζ) that bind to DNA via a leucine zipper domain.5 All members are expressed at some stage during neutrophil development, but not all at the same time.6 In particular the C/EBPs-α, -β, and -ε regulate proliferation and differentiation, and are thus essential for formation of mature neutrophils.5 C/EBP-ε is expressed as 4 isoforms of 32, 30, 27, and 14 kDa, respectively. However, only full length (ie, 32 kDa C/EBP-ε) has full transactivating potential as demonstrated in vitro.7,-9 C/EBP-ε is essential for expression of genes encoding proteins stored in specific granules of the neutrophil, such as lactoferrin and human cathelicidin antimicrobial protein (hCAP-18). In addition, C/EBP-ε induces cell cycle arrest at the myelocytic stage of differentiation and is thus important for exit from the proliferative pool at this stage of maturation.10 In accordance with this, C/EBP-ε protein expression is almost exclusively found at the myelocyte (MC)/metamyelocyte (MM) stage of differentiation.6 High amounts of C/EBP-ε messenger RNA (mRNA) are found in the MC/MM population, but a considerable amount of C/EBP-ε transcript is also detected in MBs and promyelocytes (PMs),11 indicating that the expression of C/EBP-ε may be subject to posttranscriptional regulation.
MicroRNAs (miRNAs) are increasingly recognized as important regulators of differentiation and maturation of cells through their ability to modulate protein synthesis posttranscriptionally by binding to complementary sequences of the 3′-untranslated region (UTR) of mRNAs and inhibiting translation. The functional importance of miRNAs has been demonstrated in granulopoiesis where miRNAs regulate expression of transcription factors and pathways important for correct neutrophil development.12,13 A specific miRNA signature has been associated with some AMLs indicating that miRNAs contribute to the disease phenotype in these leukemias, thus underscoring the importance of correct expression of miRNAs during normal granulocytic development.14
We have recently described the global miRNA expression pattern during human granulopoiesis, and we found microRNA-130a (miR-130a) to be highly expressed in proliferating granulocytic precursors with a steep decline in expression as the cells mature in vivo.13,15 Using miRNA target-prediction software, we identified the transcription factor C/EBP-ε as a putative target for miR-130a. The purpose of this study was to analyze the normal expression of miR-130a and C/EBP-ε in human and murine granulocytic precursors, and to investigate whether miR-130a contributes to regulation of C/EBP-ε protein levels during granulopoiesis.
Materials and methods
Isolation of neutrophils and their precursors from bone marrow and peripheral blood
Bone marrow was aspirated from the iliac crest and blood was drawn from a cubital vein of healthy volunteer donors giving informed consent. The study was approved by the local ethics committee (H-1-2011-165) and was conducted in accordance with the Declaration of Helsinki. Bone marrow cells were isolated according to their stage of maturation as previously described.11 Neutrophils were isolated from peripheral blood as previously described.6
Isolation of neutrophil precursors from murine bone marrow
Bone marrow cells were isolated from femora, tibiae and os iliae of 3 16- to 19-week-old C57BL/6 wild-type mice and 3 age-matched Cebpe−/− mice on a C57BL/6 background. The bones were crushed and homogenized in 4% fetal calf serum (FCS) in phosphate-buffered saline (PBS) using a mortar. Bone marrow cells were filtered through a 70-μm nylon cell strainer (BD Biosciences) and kept on ice. After centrifugation (4 minutes, 300 g) the cell pellet was lysed in 300 μL 10% Pharmlyse (BD Biosciences) for 1 minute and lysis was terminated by addition of 5 mL of PBS with 4% FCS. After centrifugation (4 minutes, 300 g), the supernatant was removed and the pellet was resuspended in 10 mL PBS with 4% FCS and cell number determined. Neutrophils were isolated by immunomagnetic purification with biotin-labeled mouse Anti-Ly6G antibody (130-092-332) and anti-biotin microbeads (120-003-029) on an MACS LS column (all from Miltenyi Biotec).
Cell Culture
Cells were obtained from ATCC. A549 (ATCC CCL-185) was grown in HAM F12 (Gibco BRL) supplemented with 10% FCS, (Gibco BRL), 1% 100 U/mL penicillin, and 100 μg/mL streptomycin (P/S) (Gibco BRL), HEK 293 (ATCC CRL-1573) in minimum essential media (Invitrogen), 10% FCS, 1% P/S, and 1% Glutamax (Invitrogen), NIH3T3 (CRL-1658) in Dulbecco's modified Eagle's medium (Gibco BRL), with 10% fetal bovine serum (Gibco BRL), and 1% P/S, Phoenix E (CRL-3215) in DMEM, 10% FCS, and 1% P/S, and mouse promyelocytic cell line (MPRO) (ATCC CRL-11422) in Iscove's modified Dulbecco's medium (IMDM) (Gibco BRL) with 20% horse serum (Invitrogen), 5% conditioned HM5 medium, and 1% P/S, respectively, at 37°C in a humid atmosphere with 5% CO2. Human MB/PM cells were grown in IMDM, 20% FCS, 20 ng/mL murine stem cell factor (Peprotech), 20 ng/mL recombinant human interleukin 6 (Invitrogen), and 50 ng/mL recombinant human interleukin 3 (Invitrogen), and MC/MM cells were grown in IMDM, 20% FCS, 1% P/S, and 100 ng/mL granulocyte-colony stimulating factor (G-CSF) (Neupogen [AMGEN]) at 37°C in a humid atmosphere with 5% CO2.
Cell transfection by electroporation
The transient transfection (5 × 106) MB/PM or MC/MM cells were pelleted and resuspended in 100 μL Nucleofector V solution (Amaxa Biosystems). The cells were transfected with 100 pmol locked nucleic acid (LNA)-miR-130a/negative (neg) control (Exiqon) or 20 pmol pre-miR-130a/neg control (Ambion) using AMAXA nucleoporation system, as previously described,16 and were transferred to 5 mL cell medium. MB/PMs were harvested after 4 hours and MC/MMs after 24 hours. Cell transfection and reporter enzyme assays were performed in A549 and HEK293 cells as described in Häger et al.13 Stable transfection: 5 × 106 MPRO cells were transfected with 2 μg linealized pEGP-miR-130a or pEGP-miR-NULL (Cell Biolabs) vector, respectively, as previously described using AMAXA systems according to the manufacture’s recommendations.13
Retroviral transduction
The murine C/EBP-ε coding sequence (CDS) was polymerase chain reaction (PCR)-amplified from complementary DNA clone MGC: 124002 IMAGE: 40045189, with the specific primers: 5′-GGATCCGCCACCATGTCCCACGGGACCTA-3′ and 5′-CTCGAGTCAGCTGCAGCCCCCGAC-3′. The fragment was cloned into a pMIG vector (plasmid 9044; Addgene) giving rise to pMIG-mCEBPE-CDS, which was used to transfect the Phoenix E packaging cell line17 to produce retroviral particles. Two days later, the cellular supernatant containing retrovirus was collected and filtered through a 0.45-μm filter. Cells were transduced twice using Retronectin-coated wells (CH-296; Takara) and were harvested for further analysis at 0 and 48 hours posttransduction. Cells were sorted by selection of green fluorescent protein-positive cells using a fluorescence-activated cell sorter (FACSAria III; BD Biosciences) because the transduction efficiency varied between 7% and 30%. More than 80% of the cells in the sorted population were green fluorescent protein-positive.
Differentiation of MPRO and 32Dcl3 cells
In vitro differentiation of MPRO cells (1 x105 cells/mL) was induced in medium supplemented with 10 μM all-trans retinoic acid (ATRA) and harvested on days 0 to 4 for further analysis, whereas 32Dcl3 cells were stimulated with 20 ng/mL murine G-CSF and harvested for further analysis at days 0 and 7.
Cloning of C/EBP- 3ʹ-UTR in a luciferase reporter vector
The 3′-UTR region of the C/EBP-ε mRNA was PCR-amplified with the primers 5′-ACACTAGTGGCTGGCTGGTGGATTGTG-3′ and 5′-ACAAGCTTTTTTCCCAGTCACAGTGCAACTTTAT-3′ using complementary DNA generated from RNA purified from bone marrow-derived MC/MM cells. The fragment was cloned into the pMIR-Report vector (Ambion) immediately downstream of the firefly luciferase stop codon generating the plasmid pMIR-CEBPE-wild-type (wt). The miR-130a target site in the 3′-UTR of C/EBP-ε mRNA was disrupted by mutating (mut) the miR-130a seed-match sequence using the QuickChange kit (Stratagen) and the primers 5′-CATAATGATTATATGGCTGAATAAAGAACGTGAGTGACTGGGAAAAAAGCTT-3′, 5′-GCTTTTTTCCCAGTCACTCACGTTCTTTATTCAGCCATATAATCATTATG-3′ giving rise to pMIR-CEBPE-mut. Constructs were verified by sequencing.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting
Western blot analysis was performed with antibodies against C/EBP-ε (SC-158) and β-actin (C-1212) (both Santa Cruz Biotechnology) as previously described.13
Quantitative real-time PCR
Total RNA was prepared with PureLink RNA Mini Kit (Ambion) according to the manufacturer’s recommendations and mRNA expression levels determined by quantitative real-time PCR (Applied Biosystems) as previously described15 using the following FluoroSpin 498-labeled primers: Human mRNAs: CEBPE: HS00357657_m1, LTF: HS00158924_m1, CAMP (hCAP18): Hs00189038_m1, CYBB: Hs00166163_m1, MYC: Hs00153408_m1. Murine mRNAs: Cebpe: Mm02030363_s1, Camp: Mm00438285_m1, Ltf: Mm00434797_m1, lipocalin-2 Lcn2: Mm00809552_s1, myeloperoxidase (Mpo): Mm00447875_g1, matrix metalloproteinase 9 (Mmp9): Mm 00600164_g1, Myc: Mm01192721_g1. Expression of β-actin mRNA (HEX) (human: 4326315E and mouse: 4253341E) was used for normalization. For quantification of miR-130a Taqman miRNA assay (RT/TM 454) was used and normalized to expression of SNO234 (RT/TM1234) (mouse cells) or RNU6B (RT/TM1093) (human cells).
FACS analysis: surface markers/propidium iodide/apoptosis
FACS analysis: 1 × 105 cells were labeled with biotinylated rat anti-mouse CD11b antibody (51-01712J; Miltenyi), and 1 μL streptavidin-PE (12-4317-87; eBioscience). The cells were then stained with 7-AAD (BD Pharmingen) and analyzed by FACS. Cell cycle analysis: propidium iodide staining was performed as previously described.13 The purity of bone marrow-derived cells and polymorphonuclear leukocytes (PMNs) from peripheral blood with regard to nonneutrophil cells (supplemental Table 1, available on the Blood Web site) was determined by flow cytometry as described previously.6 The fraction of megakaryocyte–erythroid progenitor cells, common myeloid progenitor cells and granulocyte-macrophage progenitor cells in the MB/PM cell population was characterized using the following surface markers: megakaryocyte–erythroid progenitor cells: Lin-, CD11b−, CD34+, CD38+, CD123−, CD45RA− (0.5% to 2%), common myeloid progenitor cells: Lin-, CD11b−, CD34+, CD38+, CD123+, CD45RA− (2% to 7%), and granulocyte-macrophage progenitor cells: Lin−, CD11b−, CD34+, CD38+, CD123+, CD45RA+ (2% to 7%).18,19
Statistical analysis
To determine significance of differences observed between the different transfections comparisons were made using the Student t test. To identify significance between expression levels of different mRNAs in human bone marrow, in differentiated MPRO, and in the reporter assays, we compared the different cell populations by one-way analysis of variance. P values <.05 were considered statistically significant.
Results
miR-130a and C/EBP-ε expression during granulopoiesis
Myeloid cells from normal human bone marrow were isolated by density centrifugation, and nonneutrophil cells were removed by immunomagnetic depletion (supplemental Table 1). This method allowed us to isolate 3 populations of neutrophil precursors representing different stages of maturation consisting largely of MBs and PMs, MCs and MMs, and band cells (BCs) and segmented neutrophils (SCs), respectively. Mature neutrophils were isolated from peripheral blood. Quantitative real-time PCR analysis confirmed high expression of miR-130a in MB/PMs and a low expression in the more mature cells confirming our previous miRNA microarray data (supplemental Figure 1A).13,15
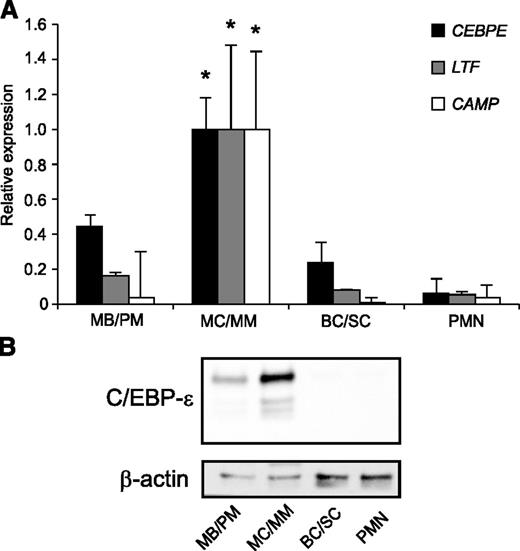
The level of C/EBP-ε mRNA was high in MB/PMs, with a further increase in the MC/MM population followed by a steep decline to low levels in BC/SCs, which remained low in PMNs (Figure 1A).11 On the other hand, C/EBP-ε protein was virtually absent in MB/PM cells, peaked in MC/MM cells with 3 different isoforms present, and almost undetectable thereafter (Figure 1B). This indicates a possible miRNA-mediated posttranscriptional regulation of C/EBP-ε protein expression. The mRNAs for the 2 specific granule proteins lactoferrin and hCAP18 both peaked in the MC/MM population in accordance with the role of C/EBP-ε as a major transcription factor for specific granule protein expression (Figure 1A).
Expression of miR-130a and C/EBP-ε during human granulopoiesis. (A) mRNA expression of CEBPE, LTF, and CAMP (hCAP18) in human MB/PM, MC/MM, BC/SC, and PMN cells measured by real-time PCR. The highest value between the 4 populations was assigned the value 1, and the relative expressions of the other samples were recalculated accordingly. Asterisks indicate significant differences between the peak expression and the other populations during granulopoiesis (*P < .05). (B) C/EBP-ε and β-actin protein expression in the 4 populations examined by western blot. Error bars indicate SD between donors.
Expression of miR-130a and C/EBP-ε during human granulopoiesis. (A) mRNA expression of CEBPE, LTF, and CAMP (hCAP18) in human MB/PM, MC/MM, BC/SC, and PMN cells measured by real-time PCR. The highest value between the 4 populations was assigned the value 1, and the relative expressions of the other samples were recalculated accordingly. Asterisks indicate significant differences between the peak expression and the other populations during granulopoiesis (*P < .05). (B) C/EBP-ε and β-actin protein expression in the 4 populations examined by western blot. Error bars indicate SD between donors.
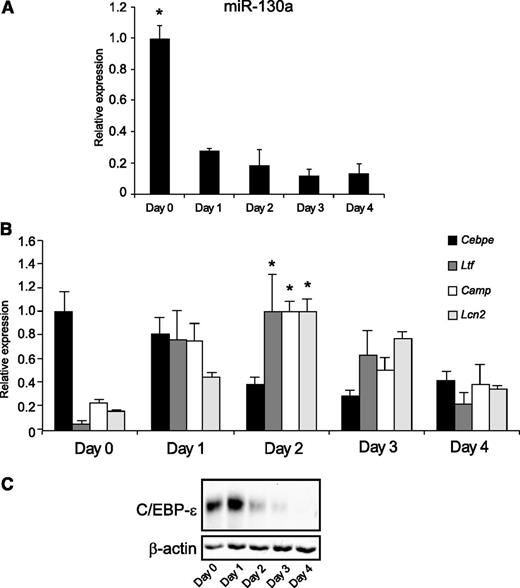
We next examined whether in vitro differentiation of the promyelocytic cell line MPRO displayed the same orderly appearance of miR-130a, C/EBP-ε, and specific granule proteins as observed during neutrophil differentiation in humans. Overall, miR-130a had the same expression pattern in differentiated MPRO cells as seen during human granulopoiesis (Figure 2A), although with a lower initial amount of miR-130a (fivefold) at day 0 cells compared with day 2 cells (the cells with peak Ltf expression) as compared with the 10-fold decline from MB/PMs to MC/MMs (Figure 1A). This might reflect the slightly more mature phenotype of MPRO cells compared with MB/PMs from bone marrow, a notion supported by the high level of C/EBP-ε mRNA at both day 0 and day 1. Transcript levels for Ltf, Camp, and Lcn2 all peaked at day 2 (Figure 2B). Western blot analysis showed high expression of C/EBP-ε protein at days 0 and 1 (Figure 2C). mRNA levels for Mpo (peak expression in PMs) and Mmp9 (peak expression in band cells) were determined and used as markers of differentiation20,21 (supplemental Figure 2B).
The miR-130a and C/EBP-ε expression in ATRA-induced MPRO cells. (A) Expression of miR-130a in differentiated MPRO cells. (B) The expression pattern for Cebpe, Ltf, Camp, and Lcn2 mRNAs in differentiated MPRO cells. The highest value of expression of each mRNA during the 5 days of differentiation was assigned the value 1, and the relative expressions between the other populations were recalculated accordingly. Asterisks indicate significant differences between the peak expression and the other days of differentiation (*P < .05). (C) Immunoblot showing expression of C/EBP-ε protein and β-actin expression in differentiated MPRO cells. Error bars represent standard deviation (SD) of triplicate measurements.
The miR-130a and C/EBP-ε expression in ATRA-induced MPRO cells. (A) Expression of miR-130a in differentiated MPRO cells. (B) The expression pattern for Cebpe, Ltf, Camp, and Lcn2 mRNAs in differentiated MPRO cells. The highest value of expression of each mRNA during the 5 days of differentiation was assigned the value 1, and the relative expressions between the other populations were recalculated accordingly. Asterisks indicate significant differences between the peak expression and the other days of differentiation (*P < .05). (C) Immunoblot showing expression of C/EBP-ε protein and β-actin expression in differentiated MPRO cells. Error bars represent standard deviation (SD) of triplicate measurements.
Target verification of miR-130a in the CEBPE 3ʹ-UTR
One theoretical miR-130a target site is present within the 3ʹ-UTR of the human C/EBP-ε mRNA (CEBPE). To investigate the effect of miR-130a on C/EBP-ε expression, we generated a reporter plasmid expressing a luciferase mRNA with the wt 3ʹ-UTR of C/EBP-ε mRNA (pMIR-CEBPE-wt), as well as a plasmid in which the miR-130a binding site in the 3′-UTR was mutated (pMIR-CEBPE-mut). Transfection of HEK293 cells, which have a low internal miR-130a level (Figure 3C) with pMIR-CEBPE-wt and pre-miR-130a caused an almost 50% reduction in luciferase activity compared with cells transfected with pre-miR-neg control. No difference in luciferase activity was observed for cells transfected with pMIR-CEBPE-mut and either pre-miR-130a or pre-miR-neg control (Figure 3A). To test the effect of inhibiting endogenous miR-130a, we used A549 cells that have a high internal miR-130a level (Figure 3C). The cells were transfected with the same 2 plasmids and either an LNA against miR-130a or a negative control LNA. Inhibition of miR-130a with a specific LNA caused a 50% upregulation of luciferase activity for the plasmid carrying the wt C/EBP-ε 3ʹ-UTR, whereas inhibition of miR-130a did not affect luciferase expression from pMIR-CEBPE-mut (Figure 3B). Together, these results show that miR-130a can regulate C/EBP-ε expression by targeting the 3ʹ-UTR of the C/EBP-ε transcript.
Target verification of miR-130a in C/EBP-ε 3ʹ-UTR. HEK293 cells or A549 cells were transfected with pMIR-CEBPE-wt or pMIR-CEBPE-mut and (A) pre-miR-130a/scrambled miRNA or (B) LNA-130a/LNA-neg, respectively. The cells were harvested 24 hours posttransfection and luciferase activity was measured using the Dual-Luciferase Reporter Assay System. The firefly luciferase activity was normalized to the activity of renilla luciferase expressed from pGL4,74 included in each transfection. Error bars show SD from triplicate measurements. The expressions from the transfections with the controls (miR-neg and LNA-neg) were assigned the value 1 and the relative expression measured from the constructs co-transfected with miR-130a or LNA-130a was recalculated accordingly. (C) Real-time PCR measuring the internal miR-130a level in A549 and HEK293 cells. Error bars indicate SD between 3 measurements. Asterisks indicate significant differences in expression between transfections (*P < .05).
Target verification of miR-130a in C/EBP-ε 3ʹ-UTR. HEK293 cells or A549 cells were transfected with pMIR-CEBPE-wt or pMIR-CEBPE-mut and (A) pre-miR-130a/scrambled miRNA or (B) LNA-130a/LNA-neg, respectively. The cells were harvested 24 hours posttransfection and luciferase activity was measured using the Dual-Luciferase Reporter Assay System. The firefly luciferase activity was normalized to the activity of renilla luciferase expressed from pGL4,74 included in each transfection. Error bars show SD from triplicate measurements. The expressions from the transfections with the controls (miR-neg and LNA-neg) were assigned the value 1 and the relative expression measured from the constructs co-transfected with miR-130a or LNA-130a was recalculated accordingly. (C) Real-time PCR measuring the internal miR-130a level in A549 and HEK293 cells. Error bars indicate SD between 3 measurements. Asterisks indicate significant differences in expression between transfections (*P < .05).
Biological effects of miR-130a overexpression in MRPO cells
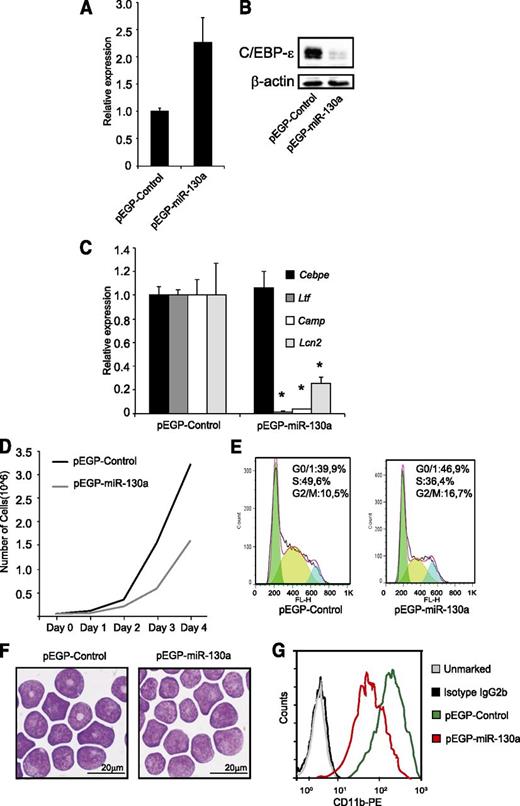
To investigate the effects of miR-130a in granulocytic precursor cells, we stably transfected MPRO cells with a plasmid expressing miR-130a or with an empty vector. Overexpression of miR-130a (Figure 4A) resulted in a significant downregulation of C/EBP-ε protein (Figure 4B). To investigate the biological effects of downregulating C/EBP-ε protein expression, we determined mRNA levels of the specific granule proteins Ltf, Camp, and Lcn2, all known to be regulated by C/EBP-ε.22 Expression of Ltf and Camp was almost absent in the pEGP-miR-130a clone compared with pEGP-control and Lcn2 expression was reduced by 75% (Figure 4C). These expression patterns mimic those seen in bone marrow–derived neutrophil precursors from the Cebpe−/− mouse (supplemental Figure 3).
The miR-130a affects cell growth and expression of mRNAs encoding specific granule protein. MPRO cells were stably transfected with the vectors pEGP-miR-130a and pEGP-miR-Null. (A) Relative miR-130a expression in the stably transfected clones. (B) Western blot analysis shows C/EBP-ε protein expression in the same cells as in (A). (C) Relative expression level of Cebpe, Ltf, Camp, and Lcn2 mRNAs in the 2 clones. Error bars indicate SD between 3 individual measurements. Asterisks indicate significant differences in expression between the 2 clones (*P < .05). (D) Cell growth of the clones over a 4-day period. (E) Cell cycle profile of the 2 clones. (F) Cytospins showing the clones stained with May-Grünwald Giemsa; scale bars = 20 μm. Cytospins were photographed using an Olympus BX51 microscope (100×/1.35 PlanApo oil objective) with an Olympus DP70 camera and the analySIS B5.0 software package (Olympus). (G) FACS analysis of the surface marker CD11b.
The miR-130a affects cell growth and expression of mRNAs encoding specific granule protein. MPRO cells were stably transfected with the vectors pEGP-miR-130a and pEGP-miR-Null. (A) Relative miR-130a expression in the stably transfected clones. (B) Western blot analysis shows C/EBP-ε protein expression in the same cells as in (A). (C) Relative expression level of Cebpe, Ltf, Camp, and Lcn2 mRNAs in the 2 clones. Error bars indicate SD between 3 individual measurements. Asterisks indicate significant differences in expression between the 2 clones (*P < .05). (D) Cell growth of the clones over a 4-day period. (E) Cell cycle profile of the 2 clones. (F) Cytospins showing the clones stained with May-Grünwald Giemsa; scale bars = 20 μm. Cytospins were photographed using an Olympus BX51 microscope (100×/1.35 PlanApo oil objective) with an Olympus DP70 camera and the analySIS B5.0 software package (Olympus). (G) FACS analysis of the surface marker CD11b.
As mentioned above, cell cycle progression is also regulated by C/EBP-ε.4,10 Targeted deletion of Cebpe is known to induce a block in differentiation at the myelocyte stage causing more cells to be retained in the proliferative stage.4 Conversely, forced expression of C/EBP-ε drives more cells into the G0/G1-phase of cell cycle, resulting in decreased proliferation and enhanced differentiation.10 We performed a growth assay and cell cycle analysis of the MPRO clones and surprisingly found a significant reduction in the rate of proliferation of the pEGP-miR-130a clone compared with the pEGP-control (Figure 4D). This was accompanied by a decline of cells in the S-phase and an accumulation of cells in the G0/G1-phase (Figure 4E), despite the reduction of C/EBP-ε. This indicates that miR-130a targets other cell cycle regulatory proteins in addition to C/EBP-ε. Furthermore, cells of the pEGP-miR-130a clone showed a more immature morphology with large round nuclei compared with cells of the pEGP-control clone of which some have a donut-shaped nucleus (Figure 4F). FACS analysis showed higher expression of the neutrophil differentiation marker CD11b23 in the pEGP-control clone than in the pEGP-miR-130a clone (Figure 4G), corroborating the more immature phenotype of MPRO cells with high internal miR-130a level.
To discriminate between the effect of miR-130a acting through C/EBP-ε and the effect of miR-130a on the expression of other cell cycle proteins, we stably transfected the myeloblast-derived cell line 32Dcl3, which does not express C/EBP-ε,24 with miR-130a. The 32Dcl3 cells that express a high level of miR-130a had reduced cell proliferation and a cell cycle profile similar to the MPRO-pEGP-miR-130a clone. Also, a more immature phenotype was seen after G-CSF-induced differentiation in the 32Dcl3 cells with high miR-130a expression (supplemental Figure 4). Together, this corroborates the findings from the MPRO-pEGP-miR-130a clone and indicates that this altered phenotype is due to miR-130a also acting on other targets than C/EBP-ε to control cell cycle and differentiation.
The aberrant phenotype of the MPRO-pEGP-miR-130a clone is partially rescued by expressing C/EBP-ε
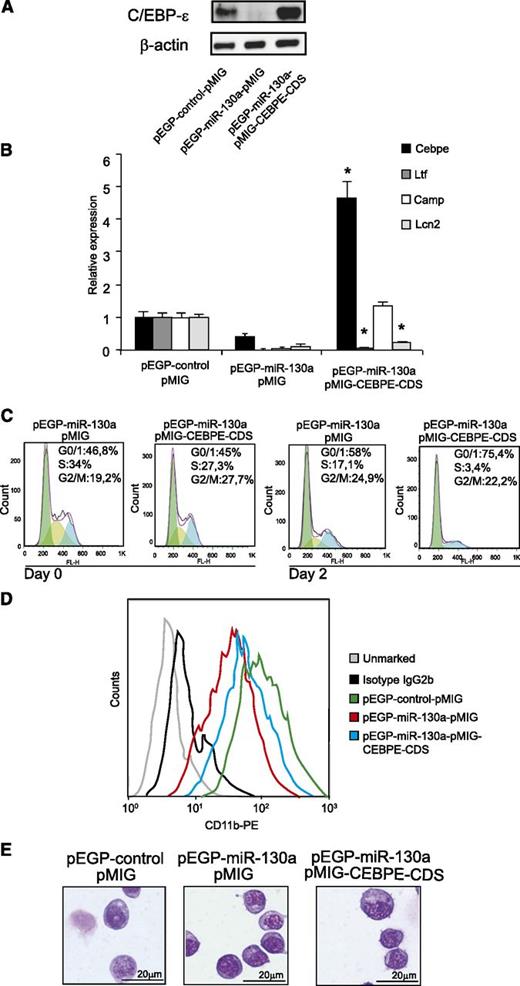
To further analyze whether the biological effects observed in MPRO-pEGP-miR-130a cells are caused by repression of C/EBP-ε synthesis, we transduced the cells with a retrovirus expressing a C/EBP-ε mRNA that was unresponsive to miR-130a (Figure 5A). As a biological readout, we investigated the transactivating activity of C/EBP-ε on the Ltf, Camp, and Lcn2 genes. As shown in Figure 5A-B, both C/EBP-ε mRNA and its corresponding protein increased in pEGP-miR-130a cells transduced with pMIG-CEBPE-CDS. Significant induction of Camp expression and a partial induction of Lcn2 were observed in pEGP-miR-130a cells transduced with pMIG-CEBPE-CDS. Surprisingly, no effect on Ltf expression was seen as a result of increased C/EBP-ε synthesis.
Expressing a C/EBP-ε mRNA unaffected by miR-130a partially restores the phenotype of the pEGP-miR-130a clone. (A) Western blot analysis showing C/EBP-ε protein expression in the pEGP-control clone after transduction with pMIG and the pEGP-miR-130a clone after transduction with either pMIG or pMIG-CEBPE-CDS. (B) Relative expression levels of Cebpe, Ltf, Camp, and Lcn2 mRNAs in the same cells as in (A) measured by real-time PCR. Error bars indicate SD between 3 measurements. (C) Cell cycle profile in the pEGP-miR-130a clone after transduction with either pMIG or pMIG-CEBPE-CDS at 0 and 48 hours posttransduction. (D) Flow analysis of surface marker CD11b. (E) Cytospins showing the pEGP-control-pMIG and the pEGP-miR-130a after transduction with either pMIG or pMIG-CEBPE-CDS stained with May-Grünwald Giemsa; scale bars = 20 μm. Photographed as in Figure 4.
Expressing a C/EBP-ε mRNA unaffected by miR-130a partially restores the phenotype of the pEGP-miR-130a clone. (A) Western blot analysis showing C/EBP-ε protein expression in the pEGP-control clone after transduction with pMIG and the pEGP-miR-130a clone after transduction with either pMIG or pMIG-CEBPE-CDS. (B) Relative expression levels of Cebpe, Ltf, Camp, and Lcn2 mRNAs in the same cells as in (A) measured by real-time PCR. Error bars indicate SD between 3 measurements. (C) Cell cycle profile in the pEGP-miR-130a clone after transduction with either pMIG or pMIG-CEBPE-CDS at 0 and 48 hours posttransduction. (D) Flow analysis of surface marker CD11b. (E) Cytospins showing the pEGP-control-pMIG and the pEGP-miR-130a after transduction with either pMIG or pMIG-CEBPE-CDS stained with May-Grünwald Giemsa; scale bars = 20 μm. Photographed as in Figure 4.
Cell cycle analysis showed that the introduction of C/EBP-ε, unresponsive to miR-130a, drives the cells of the pEGP-miR-130a clone toward cell cycle arrest, even though a large number of the cells from the pEGP-miR-130a clone are in G0/G1 phase before C/EBP-ε is introduced (Figure 5C). This demonstrates that part of the effect of miR-130a on cell cycle is mediated via its regulation of C/EBP-ε.
The pEGP-miR-130a clone transduced with pMIG-CEBPE-CDS also had a more mature phenotype compared with the pEGP-miR-130a clone as judged by CD11b expression, although the morphology of the cells seemed unchanged (Figures 5D-E).
Overexpression of miR-130a impedes ATRA-induced differentiation of MPRO cells
Next, we investigated whether a continuously high miR-130a level could suppress C/EBP-ε protein expression during ATRA-induced differentiation of MPRO cells and inhibit differentiation. In contrast to control cells, the pEGP-miR-130a clone only expressed minute amounts of C/EBP-ε protein and Ltf, Camp, and Lcn2 mRNA at day 2 of treatment with ATRA (Figures 6A-B). The cell cycle profile at day 4 after ATRA stimulation still showed a significant amount of cells in the S- and G2/M-phases in the pEGP-miR-130a clone, whereas the cells of the control clone were almost exclusively in G0/G1 phase (Figure 6C). Furthermore, cells from the pEGP-miR-130a clone showed immature morphology and low CD11b expression compared with the control clone (Figure 6D-E).
MPRO cells overexpressing miR-130a resists ATRA-induced differentiation. (A) Immunoblot of C/EBP-ε in pEGP-control and pEGP-miR-130a during stimulation with ATRA over a 5-day period. (B) The relative expression level of Ltf, Camp, and Lcn2 mRNAs at day 2 of differentiation in the clones measured by real-time PCR. Error bars indicate SD. (C) Cell cycle profile of the 2 clones at day 0 and day 4 of ATRA stimulation. (D) Cytospins showing the clones on day 4 after ATRA stimulation stained with May-Grünwald Giemsa; scale bars = 20 μm. Photographed as in Figure 4. (E) Flow analysis of surface marker CD11b. Asterisks indicate significant differences in expression between the 2 cell populations (*P < .05).
MPRO cells overexpressing miR-130a resists ATRA-induced differentiation. (A) Immunoblot of C/EBP-ε in pEGP-control and pEGP-miR-130a during stimulation with ATRA over a 5-day period. (B) The relative expression level of Ltf, Camp, and Lcn2 mRNAs at day 2 of differentiation in the clones measured by real-time PCR. Error bars indicate SD. (C) Cell cycle profile of the 2 clones at day 0 and day 4 of ATRA stimulation. (D) Cytospins showing the clones on day 4 after ATRA stimulation stained with May-Grünwald Giemsa; scale bars = 20 μm. Photographed as in Figure 4. (E) Flow analysis of surface marker CD11b. Asterisks indicate significant differences in expression between the 2 cell populations (*P < .05).
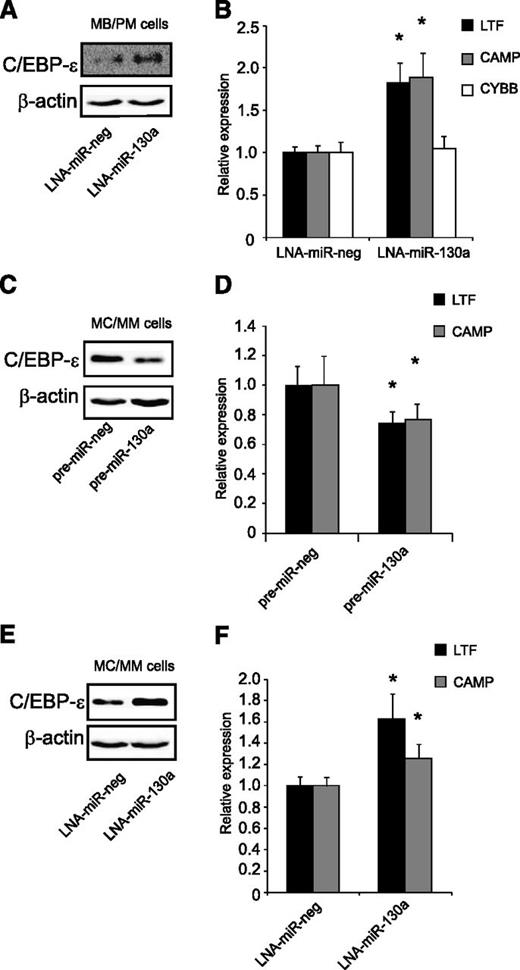
Regulation of C/EBP-ε by miR-130a in human granulocytic precursors
Immature neutrophil precursors (MB/PMs) have a high endogenous level of miR-130a and a fair amount of C/EBP-ε mRNA, but almost no C/EBP-ε protein (Figure 1). These cells were transiently transfected for 4 hours with an LNA against miR-130a (Figure 7A). Inhibition of miR-130a caused upregulation of C/EBP-ε protein, resulting in increased expression of the C/EBP-ε targets CAMP and LTF (Figure 7B). We also measured mRNA levels of cytochrome b (CYBB), which is expressed from the myelocyte stage and onward, and is expressed independent of C/EBP-ε22. The mRNA profile for CYBB showed no difference in expression between the LNA-miR-130a and the LNA-miR-neg transfected populations, indicating that the cells were at the same stage of maturity (Figure 7B).
Regulation of C/EBP-ε by miR-130a in human granulocytic precursors. (A-B) Transient transfection of MB/PM cells with LNA-miR-130a/LNA-miR-neg. The cells were harvested 4 hours after transfection. (A) Western blot analysis shows C/EBP-ε and β-actin expression in LNA-miR-neg and LNA-miR-130a transfected MB/PM cells. (B) Real-time-PCR measurements of LTF, CAMP, and CYBB. Error bars indicate SD. (C-D) MC/MM cells transiently transfected with pre-miR-130a/pre-miR-neg. The cells were harvested after 24 hours. (C) Western blot analysis examination on C/EBP-ε and β-actin. (D) Real-time-PCR measurements on LTF and CAMP. Error bars indicate SD. (E-F) MC/MM cells transiently transfected with LNA-miR-130a/LNA-miR-neg. Cell harvest after 24 hours after transfection. (E) Western blot analysis examination on C/EBP-ε and β-actin. (F) Real-time-PCR measurements on LTF and CAMP. Error bars indicate SD. The expressions from the transfections with the negative controls (pre-miR-neg and LNA-miR-neg) were assigned the value 1 and the relative expression measured from the transfected with pre-miR-130a or LNA-miR-130a was recalculated accordingly. Asterisks indicate significant differences in expression between transfections (*P < .05).
Regulation of C/EBP-ε by miR-130a in human granulocytic precursors. (A-B) Transient transfection of MB/PM cells with LNA-miR-130a/LNA-miR-neg. The cells were harvested 4 hours after transfection. (A) Western blot analysis shows C/EBP-ε and β-actin expression in LNA-miR-neg and LNA-miR-130a transfected MB/PM cells. (B) Real-time-PCR measurements of LTF, CAMP, and CYBB. Error bars indicate SD. (C-D) MC/MM cells transiently transfected with pre-miR-130a/pre-miR-neg. The cells were harvested after 24 hours. (C) Western blot analysis examination on C/EBP-ε and β-actin. (D) Real-time-PCR measurements on LTF and CAMP. Error bars indicate SD. (E-F) MC/MM cells transiently transfected with LNA-miR-130a/LNA-miR-neg. Cell harvest after 24 hours after transfection. (E) Western blot analysis examination on C/EBP-ε and β-actin. (F) Real-time-PCR measurements on LTF and CAMP. Error bars indicate SD. The expressions from the transfections with the negative controls (pre-miR-neg and LNA-miR-neg) were assigned the value 1 and the relative expression measured from the transfected with pre-miR-130a or LNA-miR-130a was recalculated accordingly. Asterisks indicate significant differences in expression between transfections (*P < .05).
Transfection of pre-miR-130a into the MC/MM population not only downregulated C/EBP-ε protein (Figure 7C), but also resulted in a significant downregulation of LTF and CAMP mRNAs (Figure 7D). In spite of the low miR-130a expression level in the MC/MM population (Figure 1A), we also transfected LNA-miR-130a into these cells to see whether their level of miR-130a expression still had a regulatory effect on C/EBP-ε protein. Western blot analysis showed a pronounced upregulation of C/EBP-ε protein in the LNA-miR-130a transfected population compared with the control (Figure 7E). This elevation of C/EBP-ε protein was accompanied by an increased expression of LTF and CAMP (Figure 7F).
Cell cycle analysis, cell morphology, and FACS analysis of the surface markers CD49d and CD11b showed no significant change between the negative control and the pre-miR-130a/LNA-miR-130a transfected cells, indicating that the transient transfections did not induce differentiation of the cells (data not shown).
Discussion
The C/EBP-ε is a transcription factor that regulates many different processes during granulopoiesis, but the mechanism(s) regulating C/EBP-ε protein expression has not yet been worked out. So far, C/EBP-ε dysfunction has only been described as a result of genetic aberrations in mice or humans,3 but in this paper, we describe a mechanism for posttranscriptional regulation of C/EBP-ε expression by miR-130a and some of the functional changes hereof.
Our results obtained both from cell lines and from normal neutrophil bone marrow precursors demonstrate that miR-130a tunes C/EBP-ε expression to a narrow peak at the myelocyte stage and that dysregulation of miR-130a contributes to altered secondary granule protein mRNA profiles, changed cell cycle regulation, and altered maturation both through regulation of C/EBP-ε, and probably also by targeting other cell cycle regulatory proteins during granulopoiesis. Expression of miR-130a is governed by c-myc25,-27 and fits the expression profiles of c-myc in neutrophil precursors from bone marrow (supplemental Figure 1B) and in ATRA-differentiated MPRO cells (supplemental Figure 2C).
Because the mRNA level of C/EBP-ε is high in human MB/PMs, one could envision that without suppression of translation by miR-130a, C/EBP-ε protein expression would peak too early during granulopoiesis. This could result in premature cell cycle arrest with decreased production of mature neutrophils and neutropenia as a consequence. The opposite situation with continuous high miR-130a levels in granulocytic precursors may also lead to a dysfunctional phenotype. This condition would generate granulocytic precursors unable to differentiate and form specific granule proteins due to downregulation of C/EBP-ε protein, thus mimicking SGD, as shown in Figures 4 and 6.
The profile of proteins stored in the different granules of the neutrophil is regulated by the temporal expression of their cognate mRNA during granulopoiesis.20 Although production of specific granule proteins is often considered to occur en bloc at the myelocyte stage of maturation, there are clear differences in their individual control of expression. Camp mRNA expression is strictly dependent on C/EBP-ε protein as seen in Figure 5, whereas mRNA expression of Lcn2 is partially induced by other transcription factors because some mRNA expression is present in the Cebpe−/− mouse (supplemental Figure 3). The Ltf expression is dependent on C/EBP-ε because neither the Cebpe−/− mouse nor patients with SGD express Ltf/LTF.28,29 It is possible, however, that miR-130a also has a direct or indirect effect on Ltf expression, independent of C/EBP-ε. This notion is supported by studies using inducible vectors with C/EBP-ε in NIH3T3 cells. Here, C/EBP-ε induces expression of Lcn2 and Mmp8 (neutrophil collagenase), but not Ltf.30 Furthermore, because the 3′-UTR of murine Ltf has a predicted target site for miR-130a, target-mediated mRNA degradation could also explain the low Ltf mRNA levels in the murine cell lines. Another possibility could be that the expression of Ltf requires another transcription factor besides C/EBP-ε, and that is also repressed by miR-130a. The fact that deprivation of C/EBP-ε does not abolish Lcn2 expression indicates that another differentiation-regulated transcription factor acts in concert with C/EBP-ε to govern the timed expression of Lcn2 in MC/MMs. In summary, our results and those of others indicates a high extent of individual regulation of specific granule protein expression.
With respect to cell cycle control by miR-130a and C/EBP-ε, we found that miR-130a inhibits proliferation of cells and that this is not fully rescued by expressing C/EBP-ε unresponsive to miR-130a. This indicates that miR-130a is implicated in cell cycle regulation of granulocytic precursors also in a manner independent of C/EBP-ε. Such a role for miR-130a has been demonstrated in prostate cancer cells (which lack C/EBP-ε) where overexpression of miR-130a significantly reduced growth of LNCaP cells.31 A role for miR-130a in cell cycle control during granulopoiesis has also been demonstrated through its inhibition of Smad4 production, although this effect is only relevant for cells induced to cell cycle arrest by transforming growth factor-β13. Predicted targets for miR-130a (using TargetScan) include the transcripts MYB,32 MYBL1,33 MDM4,34 RAB5A,35 RAB14,36 and EIF5A2,37 which all encode cell cycle promoting proteins. miR-130a was recently shown to target Dicer-1, a regulator of miRNA processing in the cytoplasm. Decreased expression of Dicer-1 by miR-130a causes downregulation of several miRNAs in the cell, which could explain the complexity of cell changes when manipulating internal levels of miR-130a in cells.38
In summary, our study reveals miR-130a as an important regulator of C/EBP-ε protein expression and thereby also as a regulator of normal granulopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors wish to thank Charlotte Horn and Tessa Hornsyld for expert technical assistance.
This work was supported by grants from The Danish Cancer Society, Lundbeck Foundation, Danish Medical Research Council, The Danielsen Foundation, The Brøchner Mortensen Foundation, and The Dana Lim Foundation.
Authorship
Contribution: M.T.L., N.B., and J.C. conceived and designed experiments; M.T.L., M.H., F.A., A.G., S.N.C., and H.M.J. performed experiments; M.T.L. analyzed data; M.T.L., M.H., A.G., and F.A. contributed reagents, materials, and analysis tools; and M.T.L., N.B., and J.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Niels Borregaard, The Granulocyte Research Laboratory, Department of Hematology 9322, National University Hospital, Blegdamsvej 9, DK-2100, Copenhagen, Denmark; e-mail: borregaard@rh.dk; or Jack B. Cowland, The Granulocyte Research Laboratory, Department of Hematology 9322, National University Hospital, Blegdamsvej 9, DK-2100, Copenhagen, Denmark; e-mail: jcowland@rh.dk.
References
Author notes
N.B. and J.B.C. contributed equally to this study.