Key Points
Egr1 haploinsufficiency in cooperation with reduced Tp53 activity accelerates the development of hematologic disease in mice.
Loss of 1 copy of Egr1 and Apc in hematopoietic stem cells, in cooperation with Tp53 loss, results in myeloid neoplasms.
An interstitial deletion of chromosome 5, del(5q), is the most common structural abnormality in primary myelodysplastic syndromes (MDS) and therapy-related myeloid neoplasms (t-MNs) after cytotoxic therapy. Loss of TP53 activity, through mutation or deletion, is highly associated with t-MNs with a del(5q). We previously demonstrated that haploinsufficiency of Egr1 and Apc, 2 genes lost in the 5q deletion, are key players in the progression of MDS with a del(5q). Using genetically engineered mice, we now show that reduction or loss of Tp53 expression, in combination with Egr1 haploinsufficiency, increased the rate of development of hematologic neoplasms and influenced the disease spectrum, but did not lead to overt myeloid leukemia, suggesting that altered function of additional gene(s) on 5q are likely required for myeloid leukemia development. Next, we demonstrated that cell intrinsic loss of Tp53 in hematopoietic stem and progenitor cells haploinsufficient for both Egr1 and Apc led to the development of acute myeloid leukemia (AML) in 17% of mice. The long latency (234-299 days) and clonal chromosomal abnormalities in the AMLs suggest that additional genetic changes may be required for full transformation. Thus, loss of Tp53 activity in cooperation with Egr1 and Apc haploinsufficiency creates an environment that is permissive for malignant transformation and the development of AML.
Introduction
Therapy-related myeloid neoplasms (t-MNs) are a late complication of cytotoxic therapy, typically for a primary malignant disease. A deletion of the long arm of chromosome 5, del(5q), or loss or deletion of chromosome 7, is frequently noted in bone marrow (BM) cells of patients with t-MN after alkylating agent therapy, implicating loss of function of tumor suppressor genes in the pathogenesis of this disease.1,-3 The identification of the involved gene(s) on chromosome 5 has been challenging, in large part because the deletions are extensive, on the order of ≥70 Mb, and homozygous deletions have not been identified.4,5 However, a number of genes and several micro RNAs (miRNAs) located on 5q, including RPS14, miRNA-145, miRNA-146a, early growth response 1 gene (EGR1), the adenomatous polyposis coli gene (APC), CTNNA1, HSPA9, and DIAPH1, have been implicated in the development of myeloid disorders caused by a gene dosage effect.3,6 Together, these studies support a haploinsufficiency model in which loss of a single allele of more than one gene on 5q act in concert to alter hematopoiesis, promote self-renewal of hematopoietic stem and progenitor cells (HSPCs), induce apoptosis of hematopoietic cells, and disrupt differentiation.
To identify leukemia-related genes on chromosome 5, we previously delineated a 970 kb commonly deleted segment (CDS) of 5q31.2 that is deleted in all patients with acute myeloid leukemia (AML) or t-MN examined.5 This is distinct from the CDS within 5q33.1 that contains RPS14, miR-145, and miRNA-146a, and is associated with myelodysplastic syndromes (MDSs) with an isolated del(5q) (the 5q syndrome).6,7 We used a candidate gene approach to identify EGR1 within the CDS of 5q31.2, as a haploinsufficient myeloid suppressor gene that cooperates with mutations induced by alkylating agents to induce myeloid leukemias in the mouse.8 In other studies, we demonstrated that APC is a critical regulator of hematopoiesis.9 APC is located at 5q22.2, which is outside the CDS, but nonetheless deleted in greater than 95% of patients with a del(5q). Moreover, we showed that Apc haploinsufficient mice (Apcdel/+) are compromised in reconstituting hematopoiesis after transplantation and develop a severe macrocytic anemia with monocytosis, reminiscent of the early stages of t-MN, that develops significantly faster when mice are also haploinsufficient for Egr1 or Tp53.10,11
Both EGR1 and APC encode proteins with tumor suppressor properties. EGR1 is a direct transcriptional regulator of many known tumor suppressor genes, including TP53, CDKN1A (p21), and PTEN,12 and has been shown to play a role in maintaining hematopoietic stem cell (HSC) quiescence and retention in the BM niche.13 APC is best known for its tumor suppressor role in colon cancer and its involvement in the WNT signaling cascade via its interaction with β-catenin.14 APC has also been found to regulate mitosis via control of spindle orientation and chromosome segregation, as well as cell migration.15
A growing body of evidence suggests that haploinsufficiency of genes on chromosome 5 cooperate with alterations of additional genes in the pathogenesis and progression of t-MN.3 Strikingly, loss of TP53 activity is strongly associated with t-MN with a del(5q) (with up to 80% of cases in some series), and the spectrum of TP53 mutations includes missense mutations in exons 4 to 8, as well as loss of the wild-type (WT) allele, typically as a result of a cytogenetic abnormality of 17p.3,16 Moreover, the results of a recent study suggest that rare preexisting clones of HSCs with TP53 mutations provide a selective growth advantage after cytotoxic therapy, and that the acquisition of additional genetic changes (ie, del[5q] or other cytogenetic abnormalities) are required for leukemic transformation.17 The TP53 tumor suppressor plays a crucial role in response to various cellular stress conditions by inducing the transcription of numerous genes controlling cell cycle arrest and apoptotic cell death.18 TP53 is also involved in the maintenance of stem cell self-renewal. Loss of Tp53 in a mouse model has been shown to promote AML by enabling aberrant self-renewal of HSCs.19,20
To model the del(5q) in mice, we used genetically engineered mice and gene knockdown strategies to examine haploinsufficiency of 2 del(5q) genes, EGR1, and APC, and of a third gene, TP53, involved in t-MN with a del(5q). Our initial studies revealed that reduction or loss of Tp53 expression in combination with Egr1 haploinsufficiency increases the rate of development of hematologic neoplasms, and influences the spectrum of diseases, suggesting cooperation between the Egr1 and Tp53 pathways. Nonetheless, none of these mice developed overt myeloid leukemia, even after administration of the alkylating agent, N-ethyl-N-nitrosourea (ENU). Next, we modeled cell intrinsic loss of Tp53 in HSPCs haploinsufficient for both Egr1 and Apc, and we demonstrated that concordant loss of these 3 genes creates an environment with genetic instability that is permissive for the development of AML in mice, thereby establishing a mouse model for t-MN with a del(5q).
Methods
Mouse strains and transplantation studies
All studies were approved by the University of Chicago Institutional Animal Care and Use Committee, and mice were housed in a fully accredited Association for Assessment and Accreditation of Laboratory Animal Care facility. Egr1+/−, Tp53−/−, and Egr1+/−, and Tp53+/− mice and corresponding littermate controls were generated by crossing Egr1+/− and Tp53+/− mice, as previously described.8,21,22 Mx1-Cre−Apcfl/+ (control mice, Apcfl/+) and Mx1-Cre+Apcfl/+ (Apcdel/+) mice were generated by crossing Apcfl/fl mice23 with Mx1-Cre transgenic mice,24 and they were injected with polyinosinic-polycytidylic acid (3 injections at 10 mg/kg GE Healthcare, Pittsburgh, PA) when mice were 2 months old, as previously described.11 In the ENU study, mice were bred on an Apcfl/+ (WT) background, and ENU was administered at 100 mg/kg, 1-month post-polyinosinic-polycytidylic acid. In the transplantation study, BM cells from Egr1+/−, Apcdel/+, and control mice were transduced with either Tp53 or Luc (control) short hairpin (shRNA) (provided by S. Lowe, Memorial Sloan-Kettering Cancer Center), and ∼3 × 106 cells were transplanted by retro-orbital injection into lethally irradiated (8.6 Gy) C57BL/6 mice.
Peripheral blood analyses and histology
A complete blood count from heart blood was determined with a Hemavet 950 counter (CDC Technologies, Oxford, CT). All organs were recovered, fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned at 4-5 μm, and stained with hematoxylin and eosin or immunostained to detect myeloperoxidase for histologic examination by a hematopathologist (J.A.). Peripheral blood, BM aspirates, and spleen touch preparations were stained with Wright-Giemsa. Spleen sections were immunostained with antibodies specific for myeloperoxidase and detected with alkaline phosphatase.
Flow cytometric analysis
Single-cell suspensions of BM and spleen were stained with fluorochrome-conjugated antibodies specific for CD71, Ter119, Gr-1, CD11b (Mac-1), CD19, immunoglobulin M (IgM), CD4, and CD8 (BD Biosciences, San Jose, CA). T-cell lymphomas were identified by the presence of massive thymic enlargement with increased volume and weight of the normal gland, and detection of the CD4 and CD8 T cell markers. Flow cytometry was performed on a FACSCanto or LSRFortessa (BD Biosciences), and data were analyzed with the FlowJo software (Tree Star, Inc., Ashland, OR).
Statistical analysis
Survival times (time to euthanasia) were estimated by the Kaplan-Meier method and compared between groups via log rank tests. A Fisher’s exact test was used to compare the incidence of disease phenotype between 2 groups (eg, percent T lymphomas in Tp53−/− vs Egr1+/−, Tp53−/− mice).
Results
Egr1 haploinsufficiency shortens disease latency and influences the spectrum of disease in Tp53+/− and Tp53−/− mice
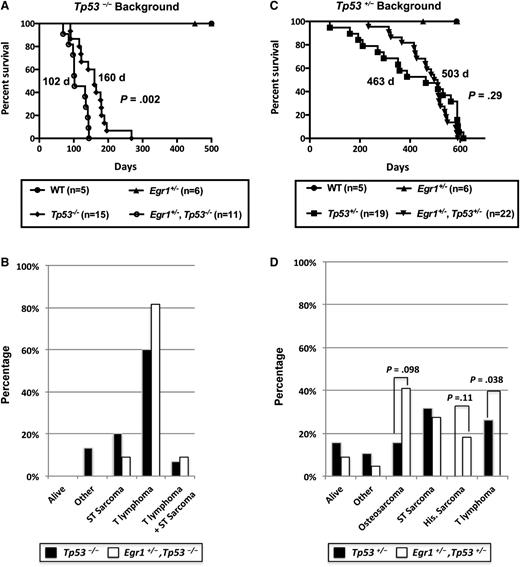
To model loss of function of Tp53 in an Egr1 haploinsufficient background, we bred Egr1+/−, Tp53−/− mice and corresponding control littermates. Egr1 haploinsufficiency significantly decreased the survival of Tp53−/− mice with a median survival of 160 days for Tp53−/− and 102 days for Egr1+/−, Tp53−/− mice (P = .002), suggesting that Egr1 haploinsufficiency cooperates with Tp53 loss in vivo to accelerate disease development (Figure 1A). Consistent with previous studies,21,22 Tp53 null mice developed both T-cell lymphomas and/or soft tissue sarcomas (Figure 1B). Loss of 1 allele of Egr1 slightly increased the incidence of T-cell lymphomas in the Tp53 null background; however, this did not reach statistical significance. The CD4 and CD8 immunophenotype of all T lymphomas is provided in supplemental Table 1, available on the Blood Web site.
Egr1 haploinsufficiency alters the disease phenotype in Tp53−/−and Tp53+/−mice. (A,C) Kaplan-Meier survival curve of Tp53−/− and Tp53+/− mice crossed with Egr1 WT or Egr1+/− mice. Percent survival (time to euthanasia of moribund animals) is plotted vs time in days. The number of mice in each cohort is shown. Egr1+/−, Tp53−/− mice had a significantly decreased survival compared with Tp53−/− mice (P = .002). The median survival for Egr1+/−, Tp53+/−, and Tp53+/− mice was not significantly different (P = .29). (B,D) Distribution of diseases developing in the mouse cohorts. All Tp53−/− crosses became moribund in <300 days. The study was terminated at ∼600 days for mice crossed on the Tp53+/− background. (B) “Other” includes 2 Tp53−/− mice that were euthanized due to a reactive condition. (D) “Other” includes 1 Tp53+/− mice that was euthanized due to severe dermatitis; 1 Tp53+/− and 1 Egr1+/−, Tp53+/− mouse that developed intestinal complications. “Osteosarcoma” includes 2 Egr1+/−, Tp53+/− mice that developed hind leg paralysis, a common symptom of osteosarcomas. P values are from a Fisher’s exact test, comparing the frequency of disease type between genotypes.
Egr1 haploinsufficiency alters the disease phenotype in Tp53−/−and Tp53+/−mice. (A,C) Kaplan-Meier survival curve of Tp53−/− and Tp53+/− mice crossed with Egr1 WT or Egr1+/− mice. Percent survival (time to euthanasia of moribund animals) is plotted vs time in days. The number of mice in each cohort is shown. Egr1+/−, Tp53−/− mice had a significantly decreased survival compared with Tp53−/− mice (P = .002). The median survival for Egr1+/−, Tp53+/−, and Tp53+/− mice was not significantly different (P = .29). (B,D) Distribution of diseases developing in the mouse cohorts. All Tp53−/− crosses became moribund in <300 days. The study was terminated at ∼600 days for mice crossed on the Tp53+/− background. (B) “Other” includes 2 Tp53−/− mice that were euthanized due to a reactive condition. (D) “Other” includes 1 Tp53+/− mice that was euthanized due to severe dermatitis; 1 Tp53+/− and 1 Egr1+/−, Tp53+/− mouse that developed intestinal complications. “Osteosarcoma” includes 2 Egr1+/−, Tp53+/− mice that developed hind leg paralysis, a common symptom of osteosarcomas. P values are from a Fisher’s exact test, comparing the frequency of disease type between genotypes.
Compared with Tp53 null mice, Tp53 heterozygous mice develop a larger spectrum of cancers at a later age, including lymphomas, soft tissue sarcomas, osteosarcomas, and more rare histiocytic sarcomas of monocyte/macrophage origin.21,25,26 To examine loss of a single allele of Tp53 (as this may occur due to chromosomal alterations) in an Egr1 haploinsufficient background, we generated Egr1+/−, Tp53+/− double heterozygous mice. Egr1 haploinsufficiency did not significantly change the overall survival of Tp53+/− mice (463 days for Tp53+/− mice vs 503 days for Egr1+/−, Tp53+/− mice; P = .29) (Figure 1C). However, there were notable differences in the frequency of diseases that developed (Figure 1D). First, there was a significant difference in the number of mice that developed lymphomas; 26% (5 of 19) of Tp53+/− mice developed T-cell lymphomas, whereas none of the Egr1+/−, Tp53+/− mice developed this disease (P = .038). Second, 18% (4 of 22) of Egr1+/−, Tp53+/− mice developed histiocytic sarcomas, with histiocytes showing varying degrees of erythrophagocytosis, whereas none of the Tp53+/− mice developed this disease in the time frame examined (P = .11). Third, osteosarcomas appeared to be more prevalent in the Egr1+/−, Tp53+/− mice, whereas only 16% (3 of 19) of Tp53+/− mice developed osteosarcomas, 41% (9 of 22) of Egr1+/−, Tp53+/−mice developed osteosarcomas or hind leg paralysis, which is a typical indicator of an osteosarcoma27 ; the latter mice were euthanized and were not examined for the presence of osteosarcomas (P = .098). Finally, an equivalent number of Tp53+/− and Egr1+/−, Tp53+/− mice developed soft tissue sarcomas (not including osteosarcomas). Together, these data infer cooperation between deregulated Egr1 and Tp53 expression during malignant progression, leading us to extend our studies to examine the role of a coordinate loss of Egr1 and Tp53, together with other genetic alterations in promoting and/or modifying disease development in vivo.
ENU accelerates and shifts tumorigenesis toward T lymphomas in Tp53+/− and Egr1+/−, Tp53+/− mice
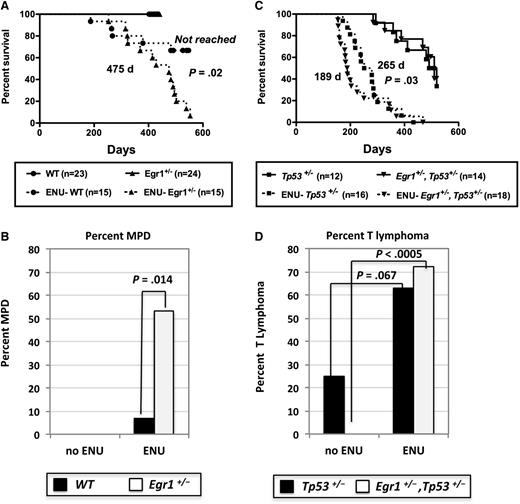
We previously showed that Egr1+/− mice do not develop disease, but when treated with the alkylating agent, ENU, almost half of the mice develop a myeloproliferative disease (MPD) with ineffective erythropoiesis.8 Using a new cohort of mice in this study, the median survival of ENU-treated Egr1+/− mice (475 days) was significantly different than ENU-treated WT mice (median not reached by 586 days; P = .02) (Figure 2A). In addition, 53% (8 of 15) of ENU-treated Egr1+/− mice developed an MPD with ineffective erythropoiesis (8 of 15; 53%) compared with only 6.7% (1 of 15) of ENU-treated WT mice (P = .014) (Figure 2B). Similar to our previously published characterization,8 mice presented with severe splenomegaly, a dramatic increase in erythropoiesis, elevated white blood cell counts, anemia, and thrombocytopenia, and atypical/dysplastic neutrophils (supplemental Table 2; data not shown). The remaining mice died predominantly of T lymphomas and soft tissue sarcomas (Table 1).
Survival curves of WT, Egr1-deficient, and Tp53-deficient mice after ENU treatment. (A,C) Kaplan-Meier survival curves of untreated and ENU-treated WT; Egr1+/−; Tp53+/−, and Egr1+/−, Tp53+/− mice. Percentage survival (time to euthanasia of moribund animals) is plotted vs time in days. The number of mice in each cohort is shown. Mice were bred on an Apcfl/+Cre− (Apc-WT) background. ENU-treated Egr1+/− mice had a significantly decreased survival compared with ENU-treated WT mice (P = .02). The survival of ENU-treated Egr1+/−, Tp53+/− mice was significantly decreased as compared with ENU-treated Tp53+/− mice (P = .03). The median survival for the Egr1+/−, Tp53+/− (505 days) and Tp53+/− (510 days) untreated cohorts was not significantly different (P = .92). (B,D) Percentage of mice that developed an MPD with ineffective erythropoiesis or a T lymphoma/T-cell leukemia in non–ENU-treated and ENU-treated mice. P values were derived using Fisher’s exact test.
Survival curves of WT, Egr1-deficient, and Tp53-deficient mice after ENU treatment. (A,C) Kaplan-Meier survival curves of untreated and ENU-treated WT; Egr1+/−; Tp53+/−, and Egr1+/−, Tp53+/− mice. Percentage survival (time to euthanasia of moribund animals) is plotted vs time in days. The number of mice in each cohort is shown. Mice were bred on an Apcfl/+Cre− (Apc-WT) background. ENU-treated Egr1+/− mice had a significantly decreased survival compared with ENU-treated WT mice (P = .02). The survival of ENU-treated Egr1+/−, Tp53+/− mice was significantly decreased as compared with ENU-treated Tp53+/− mice (P = .03). The median survival for the Egr1+/−, Tp53+/− (505 days) and Tp53+/− (510 days) untreated cohorts was not significantly different (P = .92). (B,D) Percentage of mice that developed an MPD with ineffective erythropoiesis or a T lymphoma/T-cell leukemia in non–ENU-treated and ENU-treated mice. P values were derived using Fisher’s exact test.
Comparison of outcomes of non–ENU-treated and ENU-treated mice
| . | T lymphoma or T-cell leukemia (%) . | Soft tissue sarcomas (%) . | MPD (%) . | Other (%) . | Alive (%)* . |
|---|---|---|---|---|---|
| WT | 0/18 (0) | 0/18 (0) | 0/18 (0) | 0/18 (0) | 18/18 (100) |
| Egr1+/− | 0/19 (0) | 0/19 (0) | 0/19 (0) | 0/19 (0) | 19/19 (100) |
| ENU-WT | 3/15 (20) | 1/15 (7) | 1/15 (7) | 0/15 (0) | 10/15 (67) |
| ENU-Egr1+/− | 2/15 (13) | 4/15 (27) | 8/15 (53) | 0/15 (0) | 1/15 (7) |
| Tp53+/− | 3/12 (25) | 4/12 (33) | 0/12 (0) | 0/12 (0) | 5/12 (42) |
| Egr1+/−, Tp53+/− | 0/14 (0) | 4/14 (29) | 0/14 (0) | 3/14 (21)† | 7/14 (50) |
| ENU-Tp53+/− | 10/16 (63) | 3/16 (19) | 1/16 (6) | 2/16 (12)‡ | 0/16 (0) |
| ENU-Egr1+/−, Tp53+/− | 13/18 (72) | 2/18 (11) | 1/18 (6) | 2/18 (11)§ | 0/18 (0) |
| . | T lymphoma or T-cell leukemia (%) . | Soft tissue sarcomas (%) . | MPD (%) . | Other (%) . | Alive (%)* . |
|---|---|---|---|---|---|
| WT | 0/18 (0) | 0/18 (0) | 0/18 (0) | 0/18 (0) | 18/18 (100) |
| Egr1+/− | 0/19 (0) | 0/19 (0) | 0/19 (0) | 0/19 (0) | 19/19 (100) |
| ENU-WT | 3/15 (20) | 1/15 (7) | 1/15 (7) | 0/15 (0) | 10/15 (67) |
| ENU-Egr1+/− | 2/15 (13) | 4/15 (27) | 8/15 (53) | 0/15 (0) | 1/15 (7) |
| Tp53+/− | 3/12 (25) | 4/12 (33) | 0/12 (0) | 0/12 (0) | 5/12 (42) |
| Egr1+/−, Tp53+/− | 0/14 (0) | 4/14 (29) | 0/14 (0) | 3/14 (21)† | 7/14 (50) |
| ENU-Tp53+/− | 10/16 (63) | 3/16 (19) | 1/16 (6) | 2/16 (12)‡ | 0/16 (0) |
| ENU-Egr1+/−, Tp53+/− | 13/18 (72) | 2/18 (11) | 1/18 (6) | 2/18 (11)§ | 0/18 (0) |
Mouse cohorts shown in Table 1 (and Figure 2) were bred on an Apcfl/+ (WT) background; the study was terminated at ∼500 days. The Apcfl/+(WT) mice, as with the Egr1+/+ (WT) mice, have been backcrossed to C57BL/6.
In comparison with the cohort presented in Figure 1, there were more Tp53+/− and Egr1+/−, Tp53+/− “alive” mice because the study was ended at an earlier time point. Consequently, no osteosarcomas or histiocytic sarcomas were observed because they occurred at later time points.
Two Egr1+/−, Tp53+/− mice were euthanized due to internal bleeding, and 1 died of unknown causes.
One ENU-Tp53+/− mouse was euthanized due to hind leg paralysis, and 1 developed a CD19+ IgM+ B-cell malignancy.
One ENU-Egr1+/−, Tp53+/−mouse was euthanized due to internal bleeding, and 1 developed splenomegaly with an increased proportion of CD71+ Ter119+ cells, but without anemia.
Next, we determined if secondary mutations, induced by ENU, cooperate with loss of 1 copy of Egr1 and Tp53 to accelerate or alter the MPD phenotype. When Egr1+/−, Tp53+/− mice were treated with ENU, they showed a significantly decreased survival (189 days) compared with ENU-treated Tp53+/− mice (265 days; P = .03), as well as ENU-treated Egr1+/− mice (479 days; P < .0001) (Figure 2A,C, dashed lines). In addition, Egr1+/−, Tp53+/− and Tp53+/− mice developed disease at a significantly faster rate when treated with ENU compared with non–ENU-treated mice (189 days vs 510 days; P < .0001 and 265 days vs 505 days; P < .0001, respectively) (Figure 2C). The majority of ENU-treated Egr1+/−, Tp53+/− mice developed T-cell lymphomas (13 of 18; 72%) (Figure 2D). In comparison with non–ENU-treated mice, T-cell lymphomas were increased in Tp53+/− mice (P = .067) and were significantly increased in Egr1+/−, Tp53+/− mice (P < .0005) after ENU treatment (Figure 2D). The remaining mice died predominantly of soft tissue sarcomas (Table 1). Thus, ENU treatment in Tp53+/− and Egr1+/−, Tp53+/− mice accelerates disease onset, and shifts the disease toward a T-cell phenotype.
Given that t-MN patients with a del(5q) have haploinsufficient expression of EGR1 and often have mutations and/or loss of TP53, we evaluated whether there was evidence of myelodysplasia or myeloid leukemia in mice with this combination of lesions, with or without treatment with ENU. The analysis of myeloid markers showed that none of the mice that developed T-cell lymphomas and/or soft tissue sarcomas had an accompanying myeloproliferation or myeloid malignancy (supplemental Tables 2 and 3). Many of these mice displayed splenomegaly due to panhyperplasia or erythroid hyperplasia (data not shown). However, we observed histiocytic sarcomas, a myeloid-derived neoplasm, only in Egr1+/−, Tp53+/− mice (non–ENU-treated mice). Interestingly, 1 of these mice had atypical/dysplastic megakaryocytes throughout the spleen and BM (supplemental Figure 1), raising the possibility that atypical expression of Egr1 and Tp53 may create conditions when combined with further genetic abnormalities that are conducive to the development of trilineage dysplasia, commonly seen in patients with t-MN characterized by a del(5q).
Cell intrinsic Egr1 and Apc haploinsufficiency cooperate with Tp53 loss to induce AML
As noted earlier, MDS and t-MN with a del(5q) are associated with TP53 loss or mutation, and patients with MDS with an isolated del(5q) harboring TP53 mutations have a higher risk of progressing to AML.16,28,-30 Because Tp53-null mice have a very strong preponderance to develop T-cell lymphomas, we used another approach to disrupt Tp53 function in the context of Egr1 and Apc haploinsufficiency, namely retroviral transduction–transplantation, an approach that has been used successfully to model myeloid leukemia in mice. Specifically, we used a Tp53 shRNA to knockdown expression of Tp53 in WT; Egr1+/−; Apcdel/+; and Egr1+/−, Apcdel/+ BM cells, and transplanted them into lethally irradiated C57BL/6 WT recipients (Table 2). Loss of Tp53 expression in combination with Egr1 or Apc haploinsufficiency was not sufficient for myeloid leukemia development. These mice developed only T-cell malignancies (supplemental Table 4) or soft tissue sarcomas. Transplantation of BM cells transduced with the control shRNA vector (Luc) largely did not result in disease, with the exception of 1 mouse transplanted with Egr1+/− BM that developed a soft tissue sarcoma.
Cell intrinsic Egr1 and Apc haploinsufficiency cooperate with Tp53 loss to induce AML
| BM donor . | shRNA (%) . | T lymphoma or T-cell leukemia (%) . | Soft tissue sarcoma (%) . | AML (%) . |
|---|---|---|---|---|
| WT | Luc | 0/10 (0) | 0/10 (0) | 0/10 (0) |
| Egr1+/− | Luc | 0/11 (0) | 1/11 (9) | 0/11 (0) |
| Apcdel/+ | Luc | 0/11 (0) | 0/11 (0) | 0/11 (0) |
| Egr1+/−, Apcdel/+ | Luc | 0/12 (0) | 0/12 (0) | 0/12 (0) |
| WT | Tp53 | 0/10 (0) | 1/10 (10) | 0/10 (0) |
| Egr1+/− | Tp53 | 2/10 (20) | 0/10 (0) | 0/10 (0) |
| Apcdel/+ | Tp53 | 1/11 (9) | 0/11 (0) | 0/11 (0) |
| Egr1+/−, Apcdel/+ | Tp53 | 1/12 (8) | 1/12 (8) | 2/12 (17)* |
| BM donor . | shRNA (%) . | T lymphoma or T-cell leukemia (%) . | Soft tissue sarcoma (%) . | AML (%) . |
|---|---|---|---|---|
| WT | Luc | 0/10 (0) | 0/10 (0) | 0/10 (0) |
| Egr1+/− | Luc | 0/11 (0) | 1/11 (9) | 0/11 (0) |
| Apcdel/+ | Luc | 0/11 (0) | 0/11 (0) | 0/11 (0) |
| Egr1+/−, Apcdel/+ | Luc | 0/12 (0) | 0/12 (0) | 0/12 (0) |
| WT | Tp53 | 0/10 (0) | 1/10 (10) | 0/10 (0) |
| Egr1+/− | Tp53 | 2/10 (20) | 0/10 (0) | 0/10 (0) |
| Apcdel/+ | Tp53 | 1/11 (9) | 0/11 (0) | 0/11 (0) |
| Egr1+/−, Apcdel/+ | Tp53 | 1/12 (8) | 1/12 (8) | 2/12 (17)* |
Mouse 1586 developed acute myelomonocytic leukemia; mouse 4524 developed CD19+ AML.
When Egr1+/−, Apcdel/+ double heterozygous BM cells were transduced with the Tp53 shRNA vector, 17% (2 of 12) of the mice developed AML (Table 2 and Figure 3). Both mice exhibited an elevated white blood cell count with anemia and thrombocytopenia, as well as splenomegaly, with an effacement of the normal splenic architecture (Table 3). The first mouse (1586) developed an acute myelomonocytic leukemia at 234 days, with increased monoblasts (>20%) and monocytes, with some differentiation toward granulocytes in the blood, BM, and spleen (Figure 3A). The neoplastic cells in the spleen expressed KIT, Gr-1, CD11b, and F4/80 (Figure 3B). The second mouse (4525) developed a CD19+ AML at 299 days, with >20% primitive blasts and a notable absence of mature granulocytes in the blood, BM, and spleen, suggesting there was a block in myeloid differentiation (Figure 3A). In the spleen, there was an enrichment of immature blasts that were predominantly Gr1−, KIT+, CD19+, and IgM−, and had a high mitotic rate (Figure 3B). Although a hallmark of B-cell neoplasms, CD19 has been observed in myeloid malignancies in human patients.31,32 To confirm that mouse 4525 developed an AML, we demonstrated that spleen cells were positive for myeloperoxidase, a myeloid-specific marker (Figure 3A).
The development of AML in mice transplanted with Egr1 and Apc haploinsufficient BM transduced with Tp53 shRNA. C57BL/6 recipient mice were transplanted with Egr1+/−, Apcdel/+ BM transduced with Tp53 shRNA, or Luc shRNA, as a control. Two of 12 recipient mice harboring Tp53 knockdown (green fluorescent protein-positive [GFP+]) cells, 1586 and 4525, developed AML. (A) Representative peripheral blood smears (×400), BM smears (×400), and liver section (×100) stained with hematoxylin and eosin, and spleen section (×500) immunostained with myeloperoxidase-specific antibodies are shown. Leukemic cells stain positive (pink) and lymphocytes and megakaryocytes are devoid of stain and are myeloperoxidase-negative. Images were obtained using a Zeiss Axioskop (Jena, Germany), equipped with a Zeiss Axiocam digital camera. Myeloperoxidase immunostained spleen images were obtained using an Olympus microscope (Model BX51, Tokyo, Japan) equipped with an Optromics 3CCD digital camera (Goleta, CA). Images were processed with Adobe Photoshop (San Jose, CA). (B) Flow cytometric analysis of GFP-gated spleen cells from mouse 1586 and 4525. Histograms show the expression of KIT, Gr-1, CD11b, and F4/80 in GFP-gated spleen cells from experimental leukemic (blue) and control (red) mice. (C) The percent of GFP-positive cells in the blood of mouse 1586 and 4525. The final time point is the day of euthansia. (D) The percentages of GFP-positive cells in the BM and spleen of mouse were 1586 and 4525, respectively, at the time of euthanasia. (E) Real-time polymerase chain reaction (PCR), done in triplicate, was used to assess Tp53 expression in GFP-gated BM cells before transplantation and in spleen cells from mouse 1586 and 4525, respectively. Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed relative to the control, with the standard error of the mean.
The development of AML in mice transplanted with Egr1 and Apc haploinsufficient BM transduced with Tp53 shRNA. C57BL/6 recipient mice were transplanted with Egr1+/−, Apcdel/+ BM transduced with Tp53 shRNA, or Luc shRNA, as a control. Two of 12 recipient mice harboring Tp53 knockdown (green fluorescent protein-positive [GFP+]) cells, 1586 and 4525, developed AML. (A) Representative peripheral blood smears (×400), BM smears (×400), and liver section (×100) stained with hematoxylin and eosin, and spleen section (×500) immunostained with myeloperoxidase-specific antibodies are shown. Leukemic cells stain positive (pink) and lymphocytes and megakaryocytes are devoid of stain and are myeloperoxidase-negative. Images were obtained using a Zeiss Axioskop (Jena, Germany), equipped with a Zeiss Axiocam digital camera. Myeloperoxidase immunostained spleen images were obtained using an Olympus microscope (Model BX51, Tokyo, Japan) equipped with an Optromics 3CCD digital camera (Goleta, CA). Images were processed with Adobe Photoshop (San Jose, CA). (B) Flow cytometric analysis of GFP-gated spleen cells from mouse 1586 and 4525. Histograms show the expression of KIT, Gr-1, CD11b, and F4/80 in GFP-gated spleen cells from experimental leukemic (blue) and control (red) mice. (C) The percent of GFP-positive cells in the blood of mouse 1586 and 4525. The final time point is the day of euthansia. (D) The percentages of GFP-positive cells in the BM and spleen of mouse were 1586 and 4525, respectively, at the time of euthanasia. (E) Real-time polymerase chain reaction (PCR), done in triplicate, was used to assess Tp53 expression in GFP-gated BM cells before transplantation and in spleen cells from mouse 1586 and 4525, respectively. Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed relative to the control, with the standard error of the mean.
Survival and blood counts of primary and secondary AMLs
| Mouse . | Donor cells . | Days* . | Spleen . | WBC (K/µL) . | RBC (M/µL) . | Hb (g/dL) . | PLT (K/µL) . |
|---|---|---|---|---|---|---|---|
| Controls (n = 4) | Egr1+/−, Apcdel/+ BM + Luc shRNA | 420 | 0.1 g | 9.9 ± 1.4 | 7.8 ± 0.9 | 10.6 ± 1.5 | 1377 ± 77 |
| Primary | |||||||
| 1586 | Egr1+/−, Apcdel/+ BM + Tp53 shRNA | 234 | 0.3 g | 182.4 | 3.5 | 7.4 | 572 |
| 4525 | Egr1+/−, Apcdel/+ BM + Tp53 shRNA | 299 | 0.3 g | 22.2 | 4.8 | 8.7 | 344 |
| Secondary | |||||||
| 6751 | 1586 spleen | Alive† | |||||
| 6752 | 1586 spleen | Alive | |||||
| 6753 | 1586 spleen | 111 | 1.6 g | 68.4 | 2.6 | 5.4 | 275 |
| 6754 | 1586 spleen | Alive | |||||
| 6755 | 4525 spleen | 49 | 0.4 g | 69.4 | 8.0 | 10.5 | 264 |
| 6756 | 4525 spleen | 57 | 0.2 g | 10.0 | 11.2 | 14.6 | 488 |
| 6767 | 4525 spleen | 40 | 0.3 g | 6.5 | 7.9 | 9.7 | 196 |
| Mouse . | Donor cells . | Days* . | Spleen . | WBC (K/µL) . | RBC (M/µL) . | Hb (g/dL) . | PLT (K/µL) . |
|---|---|---|---|---|---|---|---|
| Controls (n = 4) | Egr1+/−, Apcdel/+ BM + Luc shRNA | 420 | 0.1 g | 9.9 ± 1.4 | 7.8 ± 0.9 | 10.6 ± 1.5 | 1377 ± 77 |
| Primary | |||||||
| 1586 | Egr1+/−, Apcdel/+ BM + Tp53 shRNA | 234 | 0.3 g | 182.4 | 3.5 | 7.4 | 572 |
| 4525 | Egr1+/−, Apcdel/+ BM + Tp53 shRNA | 299 | 0.3 g | 22.2 | 4.8 | 8.7 | 344 |
| Secondary | |||||||
| 6751 | 1586 spleen | Alive† | |||||
| 6752 | 1586 spleen | Alive | |||||
| 6753 | 1586 spleen | 111 | 1.6 g | 68.4 | 2.6 | 5.4 | 275 |
| 6754 | 1586 spleen | Alive | |||||
| 6755 | 4525 spleen | 49 | 0.4 g | 69.4 | 8.0 | 10.5 | 264 |
| 6756 | 4525 spleen | 57 | 0.2 g | 10.0 | 11.2 | 14.6 | 488 |
| 6767 | 4525 spleen | 40 | 0.3 g | 6.5 | 7.9 | 9.7 | 196 |
Refers to the number of days posttransplant, before the mouse appeared moribund and was euthanized.
Alive refers to animals that remain disease-free at ∼170 days posttransplant.
In both mice, nonhematopoietic tissues, such as the liver, were infiltrated with malignant cells, with greater infiltration observed in mouse 1586. The leukemia was lethal when transplanted into secondary recipients. Recipient mice transplanted with 4525 spleen cells succumbed to disease quickly within 40 to 57 days. Within the time frame examined (247 days), 1 in 4 recipient mice transplanted with 1586 spleen cells developed disease within 111 days (Table 3). Secondary transplanted mice displayed a phenotype similar to the primary leukemia (Table 4). To determine whether the myeloid leukemias acquired clonal chromosomal abnormalities, we performed spectral karyotype analysis. We detected abnormal clones with structural rearrangements in both AMLs, indicating that the development of AML requires the acquisition of additional alterations. The primary AML in mouse 1586 was characterized by a t(7;17) and in mouse 4525 was characterized by a hyperdiploid karyotype and a t(6;12); related clones with evolution were noted in the secondary transplanted leukemias (Table 5, supplemental Figure 2). The karyotype of mouse 4525 lends further evidence that it developed an AML, which expresses the CD19+ marker, rather than an acute lymphoblastic leukemia. Mouse myeloid leukemias typically show hyperdiploidy with common gains of chromosomes 4, 6, 7, 8, 10, 11, 14, 15, 16, or 18, and loss of the X or Y chromosomes. The pattern of additional chromosomes in leukemia cells from mouse 4525 is consistent with that seen in myeloid leukemias (gains of 4, 6, 10, 11, 14, 15, and 16) (Table 5). To our knowledge, a mouse counterpart to human B-cell precursor acute lymphoblastic leukemia with hyperdiploidy has not been described.
Immunophenotype of primary and secondary AMLs
| BM . | Cell Type . | Control* . | 1° 1586† . | 2° 6753‡ . | 1° 4525 . | 2° 6755 . | 2° 6756 . | 2° 6757 . |
|---|---|---|---|---|---|---|---|---|
| Marker | ||||||||
| Gr1+ CD11b+ | Granulocyte/monocyte | 1.5 ± 7.9 | 50 | 27 | 0.5 | 0 | 0.5 | 1 |
| F4/80+ CD11b+ | Macrophage | 2.3 ± 0.9 | 82 | 58 | 0.8 | 0 | 0.3 | 0.4 |
| Kit+ Gr1− | Immature | 1.3 ± 0.3 | 5 | 37 | 98 | 99 | 94 | 90 |
| CD19+ IgM− | Immature B lymphoid | 1.2 ± 0.4 | 0.5 | 0.4 | 96 | 97 | 94 | 96 |
| CD19+ IgM+ | Mature B lymphoid | 4.5 ± 1.5 | 0.4 | 0.5 | 1 | 1.3 | 1.1 | 0.5 |
| CD4+ | T lymphoid | 2.8 ± 0.8 | 1 | 2.0 | 1.5 | 1.6 | 1.6 | 1.7 |
| CD8+ | T lymphoid | 0.5 ± 0.2 | 0.2 | 1.0 | 0.2 | 0.2 | 0.1 | 0.1 |
| CD71+ Ter119+ | Erythroid | 48 ± 5 | 1.5 | 0.9 | 0.3 | 0 | 0.3 | 0.4 |
| Spleen | ||||||||
| Gr1+ CD11b+ | Granulocyte/monocyte | 7 ± 2.6 | 29 | 6.0 | 1 | 0 | 0.4 | 0.5 |
| F4/80+ CD11b+ | Macrophage | 5.3 ± 1.2 | 60 | 16 | 7 | 1.0 | 0.5 | 1 |
| Kit+ Gr1− | Immature | 1.3 ± 0.3 | 20 | 67 | 82 | 98 | 89 | 88 |
| CD19+ IgM− | Immature B lymphoid | 2.3 ± 1.3 | 1 | 0.6 | 75 | 96 | 69 | 69 |
| CD19+ IgM+ | Mature B lymphoid | 12.7 ± 6.2 | 2 | 0.6 | 5 | 2.0 | 5 | 6 |
| CD4+ | T lymphoid | 6 ± 1.7 | 2 | 1.2 | 7 | 1.1 | 2.3 | 2.8 |
| CD8+ | T lymphoid | 1.3 ± 0.6 | 0.2 | 0.5 | 2 | 0.9 | 0.4 | 1 |
| CD71+ Ter119+ | Erythroid | 54.3 ± 4.1 | 6 | 6.3 | 4 | 1 | 0.5 | 0.5 |
| BM . | Cell Type . | Control* . | 1° 1586† . | 2° 6753‡ . | 1° 4525 . | 2° 6755 . | 2° 6756 . | 2° 6757 . |
|---|---|---|---|---|---|---|---|---|
| Marker | ||||||||
| Gr1+ CD11b+ | Granulocyte/monocyte | 1.5 ± 7.9 | 50 | 27 | 0.5 | 0 | 0.5 | 1 |
| F4/80+ CD11b+ | Macrophage | 2.3 ± 0.9 | 82 | 58 | 0.8 | 0 | 0.3 | 0.4 |
| Kit+ Gr1− | Immature | 1.3 ± 0.3 | 5 | 37 | 98 | 99 | 94 | 90 |
| CD19+ IgM− | Immature B lymphoid | 1.2 ± 0.4 | 0.5 | 0.4 | 96 | 97 | 94 | 96 |
| CD19+ IgM+ | Mature B lymphoid | 4.5 ± 1.5 | 0.4 | 0.5 | 1 | 1.3 | 1.1 | 0.5 |
| CD4+ | T lymphoid | 2.8 ± 0.8 | 1 | 2.0 | 1.5 | 1.6 | 1.6 | 1.7 |
| CD8+ | T lymphoid | 0.5 ± 0.2 | 0.2 | 1.0 | 0.2 | 0.2 | 0.1 | 0.1 |
| CD71+ Ter119+ | Erythroid | 48 ± 5 | 1.5 | 0.9 | 0.3 | 0 | 0.3 | 0.4 |
| Spleen | ||||||||
| Gr1+ CD11b+ | Granulocyte/monocyte | 7 ± 2.6 | 29 | 6.0 | 1 | 0 | 0.4 | 0.5 |
| F4/80+ CD11b+ | Macrophage | 5.3 ± 1.2 | 60 | 16 | 7 | 1.0 | 0.5 | 1 |
| Kit+ Gr1− | Immature | 1.3 ± 0.3 | 20 | 67 | 82 | 98 | 89 | 88 |
| CD19+ IgM− | Immature B lymphoid | 2.3 ± 1.3 | 1 | 0.6 | 75 | 96 | 69 | 69 |
| CD19+ IgM+ | Mature B lymphoid | 12.7 ± 6.2 | 2 | 0.6 | 5 | 2.0 | 5 | 6 |
| CD4+ | T lymphoid | 6 ± 1.7 | 2 | 1.2 | 7 | 1.1 | 2.3 | 2.8 |
| CD8+ | T lymphoid | 1.3 ± 0.6 | 0.2 | 0.5 | 2 | 0.9 | 0.4 | 1 |
| CD71+ Ter119+ | Erythroid | 54.3 ± 4.1 | 6 | 6.3 | 4 | 1 | 0.5 | 0.5 |
Control values determined from 3 mice transplanted with Egr1+/−, Apcdel/+ BM transduced with control Luc shRNA. The mice were euthanized between 290 to 300 days as a control.
1° is the primary mouse with AML.
2° is the secondary transplant. These mice were transplanted with leukemia cells from primary mice with AML.
Spectral karyotype analysis of primary and secondary AMLs
| Mouse . | Phenotype . | Tissue . | Karyotype (no. cells)* . |
|---|---|---|---|
| 1586 | Primary AML | Spleen | 40,XX,t(7;17)(F1;D)[8]/40,XX,der(2)t(X;2)(D;D)[1]/ 40,XX[1] |
| 6753 | Secondary AML† | Spleen | 40,XX,t(7;17)(F1;D)[7]/41,idem,del(2)(C1H3),+14[2]/ 40,idem,del(12)(BF2)[1]/ |
| 4525 | Primary AML | Spleen | 56,XX,+X,+1,+2,del(3)(BF1),+4,+4,+5,+5,+6,+6,del(6)(B3G3),t(6;12)(D;F1),+10,+11,+14,+15,+16,+17,+19[7]/58,idem,+14,+15[1]/40,XX[3]/40,XY[1] |
| 6755 | Secondary AML‡ | Spleen | 55,XX,+1,del(1)(DE3),+2,del(3)(BF1),+4,+4,+5,+5,+6,+6,del(6)(B3G3),t(6;12)(D;F1),+10,+11,+14,+15,+16,+17,+19[9] /56,idem,+X[3] |
| Mouse . | Phenotype . | Tissue . | Karyotype (no. cells)* . |
|---|---|---|---|
| 1586 | Primary AML | Spleen | 40,XX,t(7;17)(F1;D)[8]/40,XX,der(2)t(X;2)(D;D)[1]/ 40,XX[1] |
| 6753 | Secondary AML† | Spleen | 40,XX,t(7;17)(F1;D)[7]/41,idem,del(2)(C1H3),+14[2]/ 40,idem,del(12)(BF2)[1]/ |
| 4525 | Primary AML | Spleen | 56,XX,+X,+1,+2,del(3)(BF1),+4,+4,+5,+5,+6,+6,del(6)(B3G3),t(6;12)(D;F1),+10,+11,+14,+15,+16,+17,+19[7]/58,idem,+14,+15[1]/40,XX[3]/40,XY[1] |
| 6755 | Secondary AML‡ | Spleen | 55,XX,+1,del(1)(DE3),+2,del(3)(BF1),+4,+4,+5,+5,+6,+6,del(6)(B3G3),t(6;12)(D;F1),+10,+11,+14,+15,+16,+17,+19[9] /56,idem,+X[3] |
A minimum of 10 metaphase cells were analyzed.
Recipient mouse 6753 was transplanted with leukemia cells from primary mouse 1586.
Recipient mouse 6755 was transplanted with leukemia cells from primary mouse 4525.
The Tp53 shRNA construct contains GFP under the control of a SV40 promoter, and GFP can be used as a surrogate marker to estimate the proportion of the cell population with Tp53 knockdown. In the blood, there was a steady increase in GFP+ cells; approximately one-third of cells were GFP+ 50 and 130 days post-transplant in mouse 1586 and 4525, respectively (Figure 3C). When mice became moribund, GFP positivity in the blood was 40% and 67%, in the spleen it was 80% and 75%,and in the BM it was 97% and 96% for mouse 4525 and 1586, respectively (Figure 3C,D). We performed real-time PCR analysis of GFP+ sorted BM cells prior to transplantation, and spleen cells isolated from moribund mice with AML. This data revealed that the Tp53 shRNA effectively knocked down expression of Tp53 by ∼90% prior to transplantation; in AML cells, Tp53 expression was decreased by 70% to 90% (Figure 3E).
One explanation for the development of AML in only 17% of mice is that Tp53 was not knocked down in the remaining transplanted mice that did not develop myeloid neoplasms. To exclude this possibility, we used real-time PCR and showed that Tp53 expression was suppressed in most of the other mice transplanted with Egr1+/−, Apcdel/+ BM transduced with Tp53 shRNA (supplemental Figure 3). Another possible explanation for the low frequency of leukemia is that loss of the remaining Apc or Egr1 WT allele is required for disease development, as is the case for the Smad, Lkb, and Pten heterozygous knockout mice.33,-35 To examine this possibility, we compared Egr1 and Apc expression in leukemia cells from both mice to a control (Luc) transplant, and showed that these mice have similar levels of Apc expression, but slightly lower Egr1 expression. To determine whether the remaining Egr1 allele acquired a mutation affecting expression or function, we sequenced Egr1 transcripts from GFP+ sorted spleen cells from mouse 1586 and 4525; however, no mutations were found in the coding sequence of Egr1 in either mouse (data not shown).
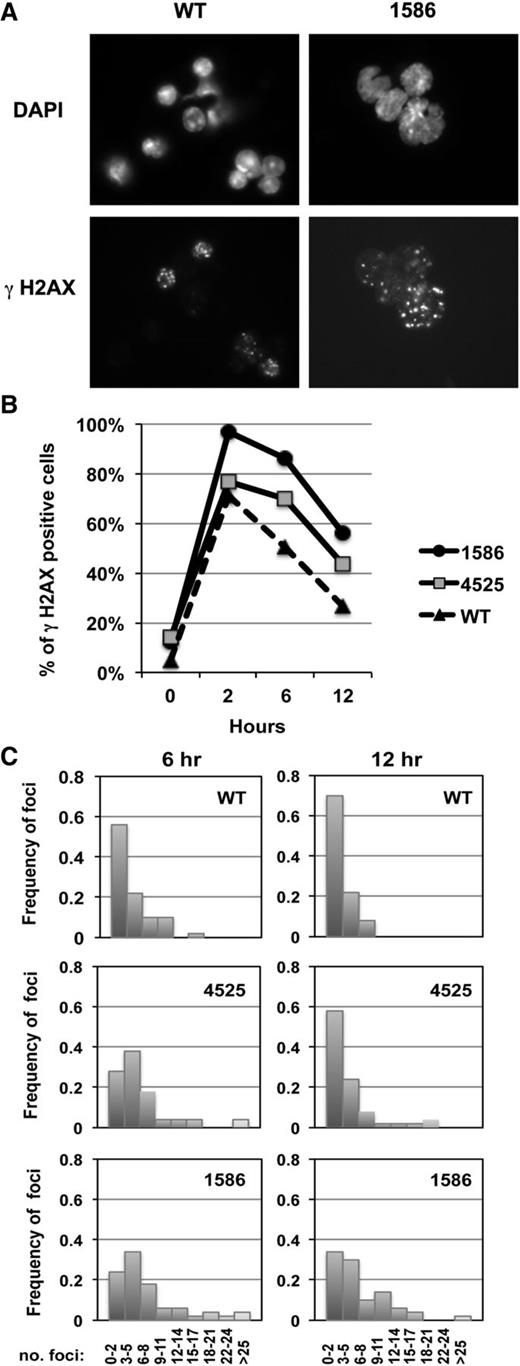
Because chromosomal deletions and translocations are the typical outcome of aberrantly repaired double-strand breaks (DSBs), the abnormal karyotype observed in the 2 AMLs suggests that the “3 hits” (ie, haploinsufficiency of Egr1 and Apc with Tp53 knock-down) create an environment that is permissive for genetic instability, leading to malignant transformation in some mice. To explore this hypothesis, we examined whether the AMLs have a differential DNA damage response to irradiation as assessed by γ-H2AX foci, a quantitative DSB biomarker (Figure 4A).36 We enumerated the percent of γ-H2AX–positive cells (Figure 4B), as well as the distribution of the number of foci (Figure 4C), from 0 to 12 hours postirradiation (2 Gy). Compared with WT control cells, both myeloid leukemias demonstrated an increase in the percentage of cells that are γ-H2AX positive at 6 and 12 hours, suggesting that both leukemias are less efficient at repairing DSBs after irradiation. Additionally, enumeration of the number of foci per cell suggested that an increased number of DSBs persist in these cells. These data, together with the structural rearrangements observed in both AMLs, support our hypothesis that concordant haploinsufficiency of Egr1 and Apc with Tp53 loss creates an environment with genetic instability.
Increased percent of γH2AX-positive cells in AMLs after irradiation. (A) Immunofluorescence microscopy of ionizing radiation-induced foci of γH2AX in irradiated splenocytes (2 Gy) isolated from a WT control and leukemic mouse transplanted with 1586 cells, 6 hours postirradiation. Nuclei were stained with 4,6 diamidino-2-phenylindole. Cells were viewed with a Zeiss Axioplan epifluorescence microscope and images were processed using Adobe Photoshop software. Original magnification, ×787.5. (B) The percentage of positive cells (≥3 γH2AX positive foci) is shown at 0 to 12 hours postirradiation in splenocytes isolated from a WT control or leukemic mice transplanted with 1586 or 4525 cells. A minimum of 100 cells were counted per time point. (C) Distribution of the number of foci per cell at 6 and 12 hours (50 cells were enumerated for each time point). An increased percentage of γH2AX-positive cells, as well as an increased number of foci per cell in AMLs compared with WT controls, is consistent with the hypothesis that DNA DSBs persist in the AMLs and contribute to genomic instability.
Increased percent of γH2AX-positive cells in AMLs after irradiation. (A) Immunofluorescence microscopy of ionizing radiation-induced foci of γH2AX in irradiated splenocytes (2 Gy) isolated from a WT control and leukemic mouse transplanted with 1586 cells, 6 hours postirradiation. Nuclei were stained with 4,6 diamidino-2-phenylindole. Cells were viewed with a Zeiss Axioplan epifluorescence microscope and images were processed using Adobe Photoshop software. Original magnification, ×787.5. (B) The percentage of positive cells (≥3 γH2AX positive foci) is shown at 0 to 12 hours postirradiation in splenocytes isolated from a WT control or leukemic mice transplanted with 1586 or 4525 cells. A minimum of 100 cells were counted per time point. (C) Distribution of the number of foci per cell at 6 and 12 hours (50 cells were enumerated for each time point). An increased percentage of γH2AX-positive cells, as well as an increased number of foci per cell in AMLs compared with WT controls, is consistent with the hypothesis that DNA DSBs persist in the AMLs and contribute to genomic instability.
Discussion
The t-MN with a del(5q) is characterized by a complex karyotype, TP53 loss or mutations, and a poor prognosis with early progression of MDS to leukemia, resistance to treatment, and short survival (median survival, 6 months).1,2 We and others have sought to identify the genes on chromosome 5 that contribute to myeloid neoplasia, and our group has previously shown that EGR1 and APC are key haploinsufficient genes on 5q.8,9,11 Recently, using a mouse model, we extended our work and demonstrated that haploinsufficiency of Egr1 and Apc cooperate, resulting in a macrocytic anemia with monocytosis and features of t-MDS.10 Herein, we build on these recent findings.
To explore the nature of cooperativity between 2 genes deregulated in t-MN patients with a del(5q) (ie, EGR1 and TP53), we used genetically engineered mice and demonstrated that reduction or loss of Tp53 expression in combination with Egr1 haploinsufficiency increases the rate of development of hematologic neoplasms and influence the spectrum of diseases. First, Egr1+/−, Tp53−/− mice and ENU-treated Egr1+/−, Tp53+/− mice developed hematologic neoplasms (ie, T-cell lymphomas at a faster pace as compared with Tp53−/− and ENU-treated Tp53+/− mice, respectively (Figures 1A and 2C). Second, Egr1+/−, Tp53+/− mice were less likely to develop T-cell lymphomas and more prone to developing histiocytic sarcomas (monocytic origin) and osteosarcomas (mesenchymal origin). Although these data suggested cooperation between the Egr1 and Tp53 pathways, none of the mice developed overt myeloid leukemia. Thus, we extended our studies to explore whether deregulated expression of an additional 5q gene, APC, was required for overt myeloid leukemia development. Herein, we establish a mouse model of t-MN with a del(5q), and show that concordant loss of Tp53 in Egr1 and Apc haploinsufficient HSPCs creates a genetically unstable environment that is permissive for the development of AML in mice.
Recent studies of the molecular pathogenesis of MDS with an isolated del(5q) (the 5q syndrome) have provided intriguing insights into the role of haploinsufficiency for cooperating genes on 5q and TP53 in the disease process. That is, TP53 activation (via stabilization) caused by haploinsufficiency for the ribosomal gene, RPS14, is the probable cause of the erythroid defect, whereas haploinsufficiency of the miR-145 and miR-146a genes may contribute to the thrombocytosis characteristic of this disease. In contrast, the acquisition of inactivating mutations of TP53 appears to play a role in progression of the disease to AML.37,38 The hypothesis that multiple genes on 5q cooperate with TP53 mutations in the pathogenesis of MDS and AML arising from MDS is supported by several recent reports of candidate gene analysis and whole genome sequencing of BM cells from MDS patients.28,-30 These studies confirm that TP53 mutations frequently co-occur with a del(5q) and tend not to occur with other gene mutations (67% of MDS patients with TP53 mutations had no point mutations in the other 93 candidate genes tested). Our study extends these findings and tests this hypothesis directly in vivo by demonstrating cooperation of del(5q) genes and aberrant Tp53 activity in a mouse model of t-MN. Although loss of Tp53 plays a role in apoptosis and genetic instability, only mice with cell-intrinsic haploinsufficient loss of both Egr1 and Apc, in addition to Tp53 knockdown, developed AML in our model. These myeloid leukemias occurred at a low penetrance and with a long latency, suggesting that additional cooperating genetic events may have been required for full transformation. Moreover, we cannot exclude a possible requirement for viral transduction, as the Tp53-specific shRNA was delivered by retroviral infection.
Chromosomal translocations are infrequently found in mouse models of AML, which typically have numerical abnormalities. At the present time, we do not know the significance of the t(7;17) or t(6;12) that we observed in the AMLs in our model. However, the observation of translocations in both AMLs strongly suggests that there was increased genetic instability and a defect in DNA repair. The increased frequency of γH2AX foci, a biomarker of DNA DSBs, after irradiation of cells from both AMLs, also supports this hypothesis (Figure 4). The chromosome 17 breakpoint region in the t(7;17)(F1;D) is syntenic to a segment of human 5q, raising the possibility that the translocation deregulates the homologs of genes on 5q. Similarly, the chromosome 6 breakpoint region in the t(6;12)(D;F1), observed in mouse 4525 with a CD19+ AML, has homology to the distal short arm of human chromosome 12, which contains several potential oncogenes and tumor suppressor genes, such as KRAS, ETV6, and CDKN1B.
In myeloid neoplasms with a del(5q), we propose that the combined EGR1 and/or APC haploinsufficiency and TP53 mutations in HSCs fosters a more penetrant phenotype, possibly due to an acquired resistance to apoptosis and increased genetic instability. Whether the recurring chromosomal abnormalities characteristic of t-MN, such as del(5q) and −7/del(7q), represent the initiating event is of considerable interest. Recent results from high-throughput genome sequencing studies shed light on this question. These studies suggest that t-MN arises from preexisting clones of HSCs with TP53 mutations that have a selective growth advantage after chemotherapy and/or radiotherapy, and that leukemic transformation requires the acquisition of additional genetic mutations.17 This process may be further influenced by modifications to the BM microenvironment, as there is extensive evidence that altered function of the BM microenvironment is also involved in disease pathogenesis.10,39,40 There is precedence for the concept that aberrant Apc and Tp53 levels cooperatively contribute to genomic instability. Precise lineage-specific doses of WNT signaling (likely disrupted in Apcdel/+ hematopoietic cells) are required for normal hematopoietic development,41 and aberrant β-catenin activity is linked to genomic instability in hematopoietic malignancies.42,-44 Most recently, the Gounari Laboratory showed that aberrant activity of β-catenin promotes genomic instability by enabling survival of thymocytes with damaged DNA, whereas compromising DSB repair, resulting in T-cell malignancies.44 Moreover, in other models with only moderately upregulated β-catenin, additional loss of Tp53 was required for lymphoma development.45 Together with our results, these data raise the possibility that the additional loss of Egr1 (a direct transcriptional regulator of Tp53) in t-MNs may be required to promote genomic instability in myeloid progenitors with aberrant Apc and Tp53 expression.
Over the past decade, several groups of investigators have sought to model myeloid neoplasms with a del(5q) by generating mice that are hemizygous for some of the putative haploinsufficient myeloid suppressor genes on 5q. However, most of these models led to aberrations in myeloid differentiation without clonal dominance, and none have led to the development of AML.3 For example, ENU-treated Egr1+/− mice developed an MPD with ineffective erythropoiesis,8 a small percentage of Diaph1+/− mice developed some myeloproliferative defects only at a very advanced age (∼450 days),46 and haploinsufficiency of the Cd74-Nid67 interval in mice, containing the Rps14 gene and modeling MDS with an isolated del(5q), caused macrocytic anemia, prominent erythroid dysplasia, and monolobulated megakaryocytes.47 Our study identifies the first genes on 5q, whereby haploinsufficiency leads to AML. We recognize that additional haploinsufficient genes on 5q, as well as alterations to the BM microenvironment, may contribute to the development of disease. Furthermore, it is possible that the critical cooperating del(5q) genes vary from patient to patient depending on additional mutations in genes, such as TP53, TET2, DNMT3A, or SRSF2. As the identification of the critical pathways leading to t-MN with a del(5q) have long been confounded by the loss of multiple genes on chromosome 5, our data provides a crucial first step in identifying therapeutic targets in this very aggressive disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zhijian Qian, Rachel Bergerson, Kenan Onel, James Downing, Scott Lowe, Scott Kogan, Kevin Shannon, David Largaespada, and Richard Larson for their helpful discussions, and Scott Lowe for the Tp53 shRNA construct.
This work was supported by National Institutes of Health, National Cancer Institute (P01 CA40046) (M.M.L.) and the Integrated Microscopy, and Cytometry and Antibody Technologies Core Facilities of the Comprehensive Cancer Center (CA14599).
Authorship
Contribution: A.S. analyzed data and wrote the manuscript; A.S., J.W., A.A.F., and E.M.D. performed experiments; J.A. analyzed histology data; T.K. assisted with the statistical analysis and reviewed the manuscript; A.S. and M.M.L.B. designed and supervised research; and M.M.L.B. cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela Stoddart, Section of Hematology/Oncology, University of Chicago, 900 E 57th St, KCBD 7th Floor, Chicago, IL 60637; e-mail: astoddar@bsd.uchicago.edu.

![Figure 3. The development of AML in mice transplanted with Egr1 and Apc haploinsufficient BM transduced with Tp53 shRNA. C57BL/6 recipient mice were transplanted with Egr1+/−, Apcdel/+ BM transduced with Tp53 shRNA, or Luc shRNA, as a control. Two of 12 recipient mice harboring Tp53 knockdown (green fluorescent protein-positive [GFP+]) cells, 1586 and 4525, developed AML. (A) Representative peripheral blood smears (×400), BM smears (×400), and liver section (×100) stained with hematoxylin and eosin, and spleen section (×500) immunostained with myeloperoxidase-specific antibodies are shown. Leukemic cells stain positive (pink) and lymphocytes and megakaryocytes are devoid of stain and are myeloperoxidase-negative. Images were obtained using a Zeiss Axioskop (Jena, Germany), equipped with a Zeiss Axiocam digital camera. Myeloperoxidase immunostained spleen images were obtained using an Olympus microscope (Model BX51, Tokyo, Japan) equipped with an Optromics 3CCD digital camera (Goleta, CA). Images were processed with Adobe Photoshop (San Jose, CA). (B) Flow cytometric analysis of GFP-gated spleen cells from mouse 1586 and 4525. Histograms show the expression of KIT, Gr-1, CD11b, and F4/80 in GFP-gated spleen cells from experimental leukemic (blue) and control (red) mice. (C) The percent of GFP-positive cells in the blood of mouse 1586 and 4525. The final time point is the day of euthansia. (D) The percentages of GFP-positive cells in the BM and spleen of mouse were 1586 and 4525, respectively, at the time of euthanasia. (E) Real-time polymerase chain reaction (PCR), done in triplicate, was used to assess Tp53 expression in GFP-gated BM cells before transplantation and in spleen cells from mouse 1586 and 4525, respectively. Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed relative to the control, with the standard error of the mean.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/7/10.1182_blood-2013-07-517953/2/m_1069f3.jpeg?Expires=1769151914&Signature=hXQR40nTeJxt7Wpf4MWa4OWtjs4b6F5NzGX1oC~IUgdGKwSup8OeiEFVpAN3J7W5G-8Z9~hECB1-gPSIHCD9XlojH8xLUx7RO61ZtZhcsV4IxEI9gBYcdpzhIFMEgepOVYhyErS6EIJ4AURzRrGhtPiXoTrqkVbne6DYgjgur5AME0-iowxOpv-97C411VPWySOlzrZLMGCVGPrImd~J72fkGTKUlDs16ESAFH~2boEM71KwcKySYLblbi8MU5R2m7C5vWGdXKKaA8smA-GfKwzaQw2dKm30O7jECgyyamQZ~~ujRb3kho48I~yJNGvJyU1i6NSSmsiCR3pzJKmWPw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)