Key Points
Mismatches in alleles C*03:03/C*03:04 were most frequent (68.7%) among the transplants with a single allele level mismatch in HLA-C.
The 7/8 C*03:03/C*03:04 mismatch group was not significantly different from the 8/8 HLA matched transplants in any transplant outcome.
Abstract
In subjects mismatched in the HLA alleles C*03:03/C*03:04 no allogeneic cytotoxic T-lymphocyte responses are detected in vitro. Hematopoietic stem cell transplantation (HSCT) with unrelated donors (UDs) showed no association between the HLA-C allele mismatches (CAMMs) and adverse outcomes; antigen mismatches at this and mismatches other HLA loci are deleterious. The absence of effect of the CAMM may have resulted from the predominance of the mismatch C*03:03/C*03:04. Patients with hematologic malignancies receiving UD HSCT matched in 8/8 and 7/8 HLA alleles were examined. Transplants mismatched in HLA-C antigens or mismatched in HLA-A, -B, or -DRB1 presented significant differences (P < .0001) in mortality (hazard ratio [HR] = 1.37, 1.30), disease-free survival (HR = 1.33, 1.27), treatment-related mortality (HR = 1.54, 1.54), and grade 3-4 acute graft-versus-host disease (HR = 1.49, 1.77) compared with the 8/8 group; transplants mismatched in other CAMMs had similar outcomes with HR ranging from 1.34 to 172 for these endpoints. The C*03:03/C*03:04 mismatched and the 8/8 matched groups had identical outcomes (HR ranging from 0.96-1.05). The previous finding that CAMMs do not associate with adverse outcomes is explained by the predominance (69%) of the mismatch C*03:03/03:04 in this group that is better tolerated than other HLA mismatches.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is an effective treatment of many hematologic disorders.1-3 Transplants in which patients and their related or unrelated donors (UDs) match in all alleles of HLA-A, -B, -C, and -DRB1 loci (8/8) have significantly superior outcomes compared with those having one or more mismatches at these loci. In the United States, 31 to 75% of patients, depending on ethnic background, are able to find an 8/8 matched UD.4,5 For patients who lack 8/8 related donors or UDs, alternative sources of allogeneic hematopoietic stem cells are HLA-mismatched UD, cord blood units, or first-degree haplo-identical relatives. Although the use of a UD with a single HLA mismatch (7/8) increases access to transplantation, transplants from HLA mismatched donors are associated with significantly higher risks for mortality and morbidity compared with those from 8/8 HLA matched donors.6-10
Several algorithms that score the risk associated with HLA mismatches based on the sharing of serologic epitopes11 and the location12 or physicochemical properties of the amino acid differences13,14 have been developed. None of these, however, has proven useful in independent validation studies. Other recent studies examining empirical outcome data identified specific HLA allele mismatch combinations or specific amino acid replacements that are associated with increased risk for severe acute graft versus host disease (aGVHD)15 or decreased survival16 compared with HLA matched transplants.
It has been proposed that the prioritization of mismatched donors could be based on an evaluation of the magnitude of relative risks for adverse outcomes.7 A Center for International Blood and Marrow Transplant Research (CIBMTR) study7 of predominately bone marrow HSCT performed using myeloablative conditioning suggested that a single HLA-A and -DRB1 mismatch appeared to be more deleterious than a single mismatch at HLA-B or -C; in contrast, a study evaluating the effect of HLA mismatches in HSCT with peripheral blood stem cells8 found higher risks for mortality in the transplants presenting one antigen mismatch in HLA-C or one mismatch in HLA-B. Both studies reported that isolated allele level mismatches in HLA-C were not associated with detrimental effects in any outcome.7,8 It has been speculated that the effect of a single mismatch at a given HLA locus is a reflection of the summation of various types of allele mismatch combinations.15,17 We reasoned that if different mismatches have distinct effects on the outcome of each individual transplant, the resulting locus effect in a cohort may depend on the prevalence of the mismatch as well as on the biological adverse impact of each mismatch. A study evaluating the cytotoxic T-lymphocyte precursor (CTLp) frequencies directed against incompatibilities at the HLA-A, -B, and -C loci in donor-recipient pairs18 showed a significant correlation between HLA class I incompatibilities and high CTLp frequencies, with the exception of pairs where the mismatch was between HLA-C*03:03/-C*03:04. In these mismatched pairs, no CTLp could be detected in spite of the fact that the alleles C*03:03/C*03:04 type distinctly by serology as HLA-Cw9 and Cw10, respectively.19
The HLA alleles C*03:03 and C*03:04 are frequently associated with the allele B*15:01 in populations with European ancestry.20 In unrelated HSCT, many donor/recipient pairs that match at HLA-B*15:01 could still have an allele level mismatch at HLA-C*03. We hypothesized that the lack of impact of the HLA-C allele level mismatches as a whole in the previous CIBMTR studies8,9 may have resulted from the high predominance and lack of immunogenicity of the HLA mismatch C*03:03/C*03:04, because this may be the most frequent allele level mismatch in HLA-C in patients of European ancestry. If this hypothesis is correct, then a negative effect of HLA-C allele level mismatches other than C*03:03/C*03:04 would have been masked.
Methods
Study population
The study included patients reported to the CIBMTR who received a transplant from a UD facilitated by the National Marrow Donor Program (NMDP) between 1988 and 2009. An initial examination of the group with an isolated HLA mismatch including the HLA alleles C*03:03/C*03:04 found that all patients had Caucasian ancestry. Therefore, the study population was restricted to Caucasian recipients to minimize other potential confounding effects. The analyses included only patients receiving their first marrow or peripheral blood stem cell UD transplantation for the treatment of acute lymphoblastic leukemia, acute myeloid leukemia, chronic myeloid leukemia, or myelodysplastic syndrome. Classification as early, intermediate, and advanced phase disease was performed as previously described.7 All surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program in accordance with the Declaration of Helsinki. Research was approved and conducted under the supervision of the NMDP Institutional Review Board. A modeling process was used, as previously described in Lee et al7 and Farag et al,21 to adjust for any bias introduced by the exclusion of nonconsenting survivors. This adjustment is standard for all studies using NMDP data.
HLA typing and HLA mismatches
High-resolution typing for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 was performed prospectively for some loci and retrospectively for other loci as previously described.7
No distinction between high-resolution/allele level and low-resolution/antigen level was made for disparities in HLA-A, -B, and -DRB1 loci. HLA-DPB1 loci were typed in a subset of patients and donors, but the impact of individual or combined mismatches in the HLA-DQB1 and -DPB1 loci on outcome was not evaluated, because prior studies in this population have not shown associations with survival. The 7/8 groups were classified into mutually exclusive categories according to the mismatches in HLA-C as follows: 1) C*03:03/C*03:04 allele level HLA mismatches (7/8 C*03:03/C*03:04MM); 2) other HLA-C allele level mismatches (involving allele mismatches with identical first 2 digits of the name other than C*03:03/C*03:04 [henceforth 7/8 C-allele≠03:03/03:04MM]); 3) C-antigen level mismatches involving alleles with differences in the first field or 2-digit name (7/8 C-antigenMM) and: 4) mismatches in alleles or antigens of HLA-A, -B, or -DRB1 (7/8 Other MM). The outcomes in these 7/8 groups were compared with those of patients matching in 8/8 alleles. In addition, the outcomes in the 7/8 group with the HLA mismatch C*03:03/C*03:04 were compared with the outcomes in other 7/8 HLA mismatched individually or combined. The transplants with 2 mismatches (6/8) were classified into 3 groups: 1) those in which one of the HLA mismatches was C*03:03/C*03:04 (6/8 C*03:03 mismatches/C*03:04MM); 2) those in which one of the mismatches occurred in HLA-C and was not C*03:03/C*03:04 (6/8 Other HLA-CMM mismatches); and 3) both mismatches occurred in loci other than HLA-C (6/8 Other MM mismatches). The outcomes in each of the 6/8 groups were compared with the outcomes in the 7/8 group in which the mismatch did not take place in HLA-C (7/8 Other MM). Directional mismatches were considered in the analysis of GVHD and engraftment, as described.6,7
Definitions of outcomes
The primary outcome was overall survival (OS), defined as time from graft infusion (day 0) to death from any cause. A number of secondary end points were also analyzed. Neutrophil engraftment was defined as achieving an absolute neutrophil count >500 × 106 cells/L by day 28 that was maintained for 3 consecutive measurements. aGVHD grade 3-4 was defined by the Glucksberg scale.22 Extensive chronic GVHD (cGVHD) was defined according to the Seattle criteria.23 Clinical relapse of the primary disease was defined by the CIBMTR criteria.24 Disease-free survival (DFS) was survival without recurrence of the primary disease. For this end point, either death or relapse was considered an event. Treatment-related mortality (TRM) was death in continuous complete remission of the primary disease.
Biostatistical methods
Probabilities for OS and DFS were calculated using the Kaplan-Meier estimator. Survival curves were compared using the log-rank test. Neutrophil engraftment was considered a dichotomous outcome and analyzed by logistic regression using a pseudo-value approach. Values for other outcomes were estimated using the cumulative incidence function.7 The point estimates of the adjusted cumulative incidence rates were based on proportional subdistribution hazards models.25-27 The adjusted cumulative incidences were compared at 100 days after transplantation for acute GVHD grade 3-4, at 5 years after transplantation for TRM, and at 2 years for cGVHD. Death was considered a competing risk for all of the end points except OS and DFS. Relapse was also considered a competing event for TRM. Patients were censored when they underwent a second HSCT procedure or, if alive, at last follow up.
To analyze the association between the type of HLA mismatch and clinical outcomes with an adjustment for other clinical risk factors, multivariate proportional hazards models were created that allowed pairs mismatched at the HLA-C antigen or allele level to be compared with each other, with HLA mismatches at other loci, and with HLA matched pairs. Models included any clinical factors that were related to a given outcome at P ≤ .05. Potential clinical covariates included disease, disease stage, Karnofsky performance status, donor-patient cytomegalovirus serostatus match, patient age, T-cell depletion, use of total body irradiation, graft source (peripheral blood or bone marrow), donor age, patient-donor sex match, and year of transplantation. All variables were tested for affirmation of the proportional hazards assumption. Variables that did not satisfy the proportional hazards assumption were adjusted for by stratification. A stepwise model-building procedure was used to develop models for each outcome with a threshold of ≤0.05 for both entry and retention in the model. Multivariate models for OS, DFS, relapse, TRM, aGVHD, and cGVHD using the Cox proportional hazards model were built. No significant interactions were identified between HLA matching and the adjusted clinical factors at P ≤ .01. Center effect was tested and was not present. Because of multiple testing, a significant P value was considered ≤.01. No additional subanalyses, including stratification of patients according to disease stage, were performed due to limiting sample sizes in the primary HLA mismatched groups of interest (7/8 C*03:03/C*03:04MM and 7/8 C-allele≠03:03/03:04MM).
Results
Frequencies of fully HLA matched and different types of single HLA mismatch transplants
A total of 7349 transplants from UDs performed between 1988 and 2009 were evaluated. This study included 4779 pairs matched in 8/8 alleles and 1854 matched in 7/8 alleles (Table 1); 716 transplants matched in 6/8 alleles (supplemental Table 1, available on the Blood Web site) were also examined.
C*03:03 or C*03:04 were found in 24.6% of the patients receiving a transplant from an 8/8 (23.4%) or a 7/8 (27.8%) matched donor. B*15:01 was present in 12.4% of the patients receiving 8/8 and 7/8 matched transplants; in contrast, and as expected from the known associations,28 B*15:01 was present in 94.0% of the patients receiving a 7/8 transplant with the mismatch C*03:03/C*03:04. Among the patients transplanted with a 7/8 donor with a mismatch other than C*03:03/C*03:04 (n = 1720), 8.3% carried B*15:01 and C*03:03 or C*03:04.
Table 1 shows the population characteristics of 8/8 and 7/8 HLA matched groups according to the type of mismatch in HLA-A, -B, -C, and -DRB1 loci. The HLA mismatch C*03:03/C*03:04 (n = 134) was present in 7.2% of the 7/8 transplants. Mismatches in alleles C*03:03/C*03:04 were most frequent (n = 134, 68.7%) among the transplants with a single allele level mismatch in HLA-C (n = 195). The 8/8 group included more transplants performed in 2005 to 2009 and were more likely to be matched at HLA-DQB1. The transplants with the HLA mismatch C*03:03/C*03:04 had the lowest level of matching in HLA-DPB1.
Single locus mismatches
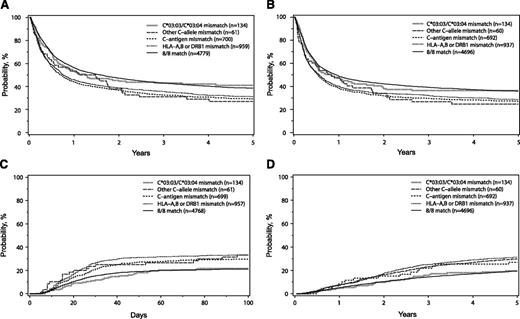
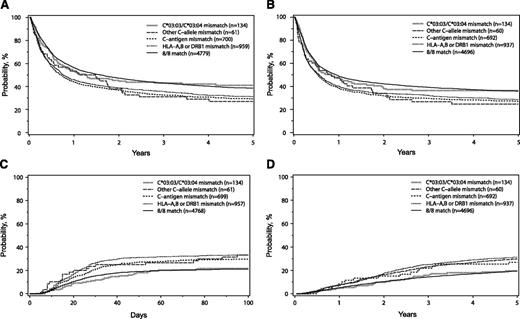
The 7/8 C*03:03/C*03:04MM group was not significantly different from the 8/8 HLA matched transplants in any transplant outcome. Indeed, the hazard ratios for survival, DFS, TRM, grade 2-4, and 3-4 aGVHD in the C*03:03/C*03:04 mismatched transplants compared with the 8/8 group suggest that these outcomes were essentially identical, all with P values > .73 (Table 2). The 8/8 matched group and the 7/8 C*03:03/C*03:04MM group had virtually identical adjusted OS (Figure 1A) and DFS (Figure 1B) 5 years after transplantation; similarly, these groups also had comparable incidence of grade 3-4 aGVHD (Figure 1C) 100 days after transplantation and TRM (Figure 1D) 5 years after transplantation. The adjusted probabilities for the 8/8 and 7/8 C*03:03/C*03:04MM groups for OS (39%, confidence interval [CI] 37%-40%; and 41%, CI 33%-49%, respectively) and DFS (36%, CI 34%-37%; and 36%, CI 29%-44%, respectively) 5 years after transplantation were virtually identical; similarly, these groups showed similar incidences of grades 3-4 aGVHD 100 days after transplantation (21%, CI 20%-22%; and 22%, CI 15%-28%, respectively). The adjusted probabilities for TRM 5 years after transplantation in the 8/8 (36%, CI 35%-38%) and 7/8 C*03:03/C*03:04MM (35%, CI 27%-43%) groups also were not significantly different (supplemental Table 2).
Adjusted probabilities of OS (A), DFS (B), grade 3-4 aGVHD (C), and TRM (D) in patients presenting no mismatch (8/8), HLA-C*03:03/C*03:04 mismatch, other HLA-C allele mismatch, HLA-C antigen mismatch, or one mismatch in the HLA-A, -B, or -DRB1 loci.
Adjusted probabilities of OS (A), DFS (B), grade 3-4 aGVHD (C), and TRM (D) in patients presenting no mismatch (8/8), HLA-C*03:03/C*03:04 mismatch, other HLA-C allele mismatch, HLA-C antigen mismatch, or one mismatch in the HLA-A, -B, or -DRB1 loci.
In contrast, mismatching at a single antigen of HLA-C (7/8 C-antigenMM) and a single allele or antigen mismatch of HLA-A, -B, or -DRB1 (7/8 Other MM) was associated with lower OS and DFS as well as with higher TRM and aGVHD grade 3-4 compared with the 8/8 HLA matched pairs (Table 2; Figure 1). The adjusted probabilities for OS 5 years after transplantation in the 7/8 C-antigen MM (29%, CI 26% to 32%) and the 7/8 Other MM (31%, CI 28% to 34%) groups were lower than those observed for the 7/8 C*03:03/C*03:04MM (P = .007 and 0.02, nonsignificant, respectively) (supplemental Table 2) and the 8/8 (P < .0001 for both comparison) (supplemental Table 2) groups. TRM and aGVHD 3-4 were significantly higher in the 7/8 Other MM group compared with the C*03:03/C*03:04MM group (Table 3). Similarly, the 7/8 C-antigen MM group presented significantly higher incidence of TRM and worse OS than the C*0303/C*0304MM (Table 3).
The 7/8 C-allele≠03:03/03:04MM had a small sample size (n = 61), which may have limited the power to detect differences. In spite of this limitation, grade 2-4 aGVHD was significantly higher in this group (hazard ratio [HR] = 1.56 and P = .0096) compared with the 8/8 group (Table 2). The 7/8 C-allele≠03:03/03:04MM also had lower OS adjusted probabilities (27%, CI 16%-38%) than the 8/8 and 7/8 C*03:03/C*03:04MM groups, although these differences were not statistically significant (Tables 2 and 3). The point estimates in the 7/8 C-allele≠03:03/03:04MM and the 7/8 antigen level mismatched groups presented comparable adjusted probabilities for DFS 5 years after transplantation (ranging from 25% to 29%), grade 3-4 aGVHD at day 100 (ranging from 30% to 33%), and TRM 5 years after transplantation (ranging from 47% to 51%) (supplemental Table 2).
The mismatch C*03:03/C*03:04 was also scored in the host-versus-graft or graft-versus-host vector; there were no significant differences in any of the transplant outcomes for this pairwise comparison or for the individual comparison with the 8/8 matched transplants (data not shown).
There were no significant differences between any of the 7/8 and 8/8 matched pairs for neutrophil engraftment and cGVHD. Table 2 also shows that there was no reduction in relapse or an enhanced graft-versus-leukemia effect conferred by any of the single HLA mismatch groups, an observation made in recent studies.6-8,10
Outcomes in groups with 2 HLA mismatches
The transplants with 2 mismatches at HLA-A, -B, -C, and -DRB1 loci were classified into 3 mutually exclusive groups according to the occurrence of one mismatch in the alleles C*03:03/C*03:04 (n = 60), the presence of one mismatch in HLA-C other than C*03:03/C*03:04 (n = 550), or in which none of the mismatches occurred in HLA-C (n = 106). The outcomes in each of these groups were compared with those in 7/8 transplants presenting a single antigen or allele mismatch in HLA-A, -B, or -DRB1 loci (n = 959). These comparisons were designed to evaluate the effect of a specific second HLA mismatch. Groups differed in the year of transplantation (P < .0001), graft type used (P < .0001), conditioning regimen (P = .003), GvHD prophylaxis (< .0001), patient age (P < .0001), and diagnosis (P = .0009).
Transplants matched at 6/8 alleles where at least one mismatch occurred in HLA-C (6/8 Other HLA-CMM) and was not C*03:03/C*03:04 had significantly worse survival (HR = 1.38; P < .0001), DFS (HR = 1.33; P < .0001), and TRM (HR = 1.35; P = .0001) compared with the 7/8 matched group (supplemental Table 3). The transplants with 2 or more mismatches that included a mismatch at C*03:03/C*03:04 or those including mismatches only in HLA-A, -B, or -DRB1 (no C mismatch) showed no significant differences in any outcome when compared with the 7/8 reference group, although with HRs that suggest risk for adverse outcomes for OS (1.29 and 1.21, respectively), DFS (1.27 and 1.16, respectively), and TRM (1.17 and 1.20, respectively).
Discussion
In spite of the higher prevalence of mismatches at HLA-DQ and -DP loci, we could not detect different outcomes between transplants mismatched only in the HLA alleles C*03:03/C*03:04 and the fully 8/8 HLA matched groups. The sample size of the C*03:03/C*03:04 mismatched group and the point estimates for mortality, DFS, and grade 3-4 aGVHD suggest that the failure to see significant differences compared with the 8/8 matched group was not due to limited power. In contrast, higher risks were seen with single mismatches at other loci and with C antigen mismatches. These results suggest that mismatches in the HLA alleles C*03:03/C*03:04 are better tolerated than other HLA mismatches where higher risks of aGVHD and lower survival are readily detectable. Certain transplant protocols only allow for the selection of fully HLA matched donors; the findings from the present study strongly suggest that the transplant eligibility criteria could be extended to patients that have a donor mismatched only in C*03:03/C*03:04.
In the present study, patient characteristics, conditioning, and graft choices varied significantly between the groups with different match grades and mismatch types. The transplants performed in recent years had higher HLA match grades on average. These differences may have resulted from improvements in HLA typing and resolution, paired with the implementation of more stringent donor selection criteria following studies that demonstrated the adverse effect of HLA mismatching; similarly, patient eligibility criteria, treatment options, and therapies have changed over time as well. In the present study, appropriate adjustments for the different factors were made when assessing outcomes in groups with different types of mismatches. The present study is in agreement with previous studies6,7 that showed that the 6/8 transplants had a significantly worse outcome than the 7/8 transplants; because of power limitations and significant differences in the time periods of performance of transplants matched in 6/8 and 7/8 loci, we cannot conclude whether the addition of the mismatch in the HLA alleles C*03:03/C*03:04 to another mismatch in HLA-A, -B, or -DRB1 loci has either a negligible or deleterious effect in transplant outcome.
The HLA alleles C*03:03 and C*03:04 differ by a single amino acid substitution at residue 91. The initial crystallographic analyses of HLA class I molecules29 showed that this residue is located in a loop connecting the second α helix of the α-1 domain with the first β-pleated sheet of the α-2 domain; residue 91 is a contact site with neither peptide30 nor with the T-cell receptor.31 A study conducted by Oudshoorn and co-workers18 suggested that the mismatch in the HLA alleles C*03:03/C*03:04 alleles cannot elicit direct T-cell allo-recognition. These alleles may have equivalent peptide presentation properties; the inconsequential effect of this mismatch in transplant outcomes may be a reflection of their functional similarities. In contrast, substitutions affecting peptide binding appear to significantly affect T-cell allo-recognition.32 Transplants in which the patient and donor present HLA mismatches in alleles that differ by substitutions at these residues presented increased risk for acute and cGVHD and death.33
Among the common single mismatches found in the present and other studies7,34,35 in 7/8 matched transplants, the mismatch in the alleles C*03:03/C*03:04 is unique in terms of the location of the structural differences; in contrast, virtually all other common mismatches differ by at least one amino acid substitution at residues that affect peptide binding35 (data not shown from the present study). Therefore, in the evaluation of HLA mismatches, the examination of the location of the distinguishing substitutions being either contact or noncontact with the peptides bound appears be a useful tool for prioritization of donors.
HLA C*03:03 and C*03:04 associate with B*15:01 in populations with European ancestry.20,28 In the present study, 7.2% of all 7/8 transplants presented the mismatch C*03:03/C*03:04; the majority of the patients with this mismatch (94.0%) carried B*15:01. The alleles B*15:01 and C*03:03 or C*03:04 were present in 14.4% of the patients receiving a transplant with a 7/8 matched donor. Although the present study is a retrospective examination of transplants and the criteria used for selection of donors are unknown, we estimate that a significant proportion (ranging from 7.2% to 14.4%) of patients with European ancestry that only have a choice for a 7/8 matched donor may benefit from selection of a C*03:03/C*03:04 mismatch.
The examination of the ethnic background of transplants presenting the HLA mismatch C*03:03/C*03:04 showed that all patients had European ancestry; this was anticipated, because these 2 structurally related alleles have similar frequencies and associate with the same allele at a contiguous locus. The isolated HLA mismatch C*03:03/C*03:04 is found often in Europeans, because both alleles associate almost equally with B*15:01 (ratio = 3:2, extracted from Maiers et al28 ). In Asians, C*03:03/C*03:04 associates with B*40:02, but at more unequal frequencies (ratio = 1:4, extracted from Maiers et al28 ). If putatively well-tolerated mismatches can be defined on the basis of the location of the residues distinguishing mismatched alleles, it can be anticipated that in other ethnic groups, other putatively permissive mismatches also may be found relatively often. For example, the HLA allele B*35:04 differs from B*35:09 and B*35:12 by single replacements at residues 131 and 103, respectively, that are also located at connecting loops. Therefore, these mismatches could potentially be permissible and could benefit patients of South, Central, or Meso American descent where these HLA-B35 alleles are found associated tightly with the same allele at the HLA-C locus (C*04:01).36
It is conceivable that after allogeneic HSCT, the mismatch in the HLA alleles C*03:03/C*03:04 may elicit allo-reactive T-cell responses through indirect recognition of C*03-derived peptides complexed with major histocompatibility complex–heterodimer molecules in the patient’s tissues. In this scenario, the recognition of HLA-derived peptides would resemble that of a minor histocompatibility antigen with similar impact in transplant outcomes.37-40 Although the present study indicates that the single mismatch in the HLA alleles C*03:03/C*03:04 has no effect on outcome, this mismatch could be significant if combined with mismatches in other loci in a similar manner as the effect observed for mismatches in the low expression HLA loci -DRB3/4/5, -DQ, and -DP,41 as well as for the presence of adverse outcome factors17 that have weak or no effect when evaluated individually.
The results described here confirm that the allele pair C*03:03/C*03:04 is the most frequent allele level mismatch in HLA-C in this Caucasian cohort. Although the differential serologic recognition of Cw9 (serotype of C*03:03) and Cw10 (serotype of C*03:04) by allo-antibodies was identified several decades ago,42 no studies have been conducted to examine whether humoral responses may be mounted following allo-recognition of epitope differences in donor/recipient pairs mismatched only in these subtypes of HLA-C*03. A recent study (M.A.F.-V. and Dolly Tyan, Stanford University, written communication) evaluated sera from 93 solid organ transplant candidates that displayed reactivity against C*03:03, C*03:04, or both alleles. Most sera reacted simultaneously with both subtypes of C*03, whereas sera from only 5 patients (5.4%) reacted only with one and not with the other subtype of C*03. Interestingly, neither of the patients whose sera reacted with C*03 carried an allele of the C*03 group. Because of the low incidence of sera with such differential reactivity, it can be speculated that the serologic epitopes distinguishing C*03:03/C*03:04 have a low immunogenicity score. In the present study, the C-allele level mismatch C*03:03/C*03:04 did not associate with increased risk for neutrophil engraftment; this observation is consistent with the hypothesis that the mismatch C*03:03/C*03:04 is not able to elicit a strong cellular or humoral allo-reactive response.
The present study provides evidence indicating that the mismatch C*03:03/C*03:04 is better tolerated and results in superior outcomes compared with other single HLA mismatches. The apparent lack of effect of HLA-C allele level mismatch in the study conducted by Lee and co-workers7 most likely resulted from the predominance of this single, nonimmunogenic mismatch. Because other allele level mismatches at HLA-C may confer higher risk for adverse outcomes, high resolution HLA-C typing is warranted for evaluation of donor/recipient allele level match grade. Previous studies6-8,43 did not show significant differences in outcomes between transplants performed with mismatches in HLA-A, -B, or -DRB1 loci or antigen level mismatches in HLA-C; the findings in the present study strongly suggest that mismatches in HLA-C alleles other than the mismatch C*03:03/C*03:04 result in inferior outcomes. With the exception of the permissible mismatch C*03:03/C*03:04, all mismatches in HLA-A, -B, -C, and -DRB1 appear to be equivalently detrimental. For patients carrying C*03 alleles, donors mismatched for C*03:03/C*03:04 should be given priority over donors with other types of HLA mismatches. Investigation and evaluation of structural differences between alleles differing only in residues that are not contact sites with peptides may lead to the identification of other permissible mismatches, with practical implications in improving the outcomes of UD transplants by optimizing the criteria for donor selection or expanding the pool of acceptable UD. The criteria of mismatch permissibility described here may rationally be extended to other graft sources of allogeneic hematopoietic stem cells as well as to organ transplantation, and these studies should be performed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute, the National Heart, Lung, and Blood Institute, and the National Institute of Allergy and Infectious Diseases, Grant/Cooperative Agreement 5U01HL069294 from National Heart, Lung, and Blood Institute and National Cancer Institute, contract HHSH234200637015C with the Health Resources and Services Administration, grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research, and grants from Allos Inc., Amgen Inc., Angioblast, an anonymous donation to the Medical College of Wisconsin, Ariad, Be the Match Foundation, Blue Cross and Blue Shield Association, Buchanan Family Foundation, CaridianBCT Celgene Corporation, CellGenix GmbH, Children’s Leukemia Research Association, Fresenius-Biotech North America Inc., Gamida Cell Teva Joint Venture Ltd, Genentech Inc., Genzyme Corporation, GlaxoSmithKline, HistoGenetics Inc., Kiadis Pharma The Leukemia & Lymphoma Society, The Medical College of Wisconsin, Merck & Co. Inc., Millennium: The Takeda Oncology Co., Milliman USA Inc., Miltenyi Biotec Inc., National Marrow Donor Program, Optum Healthcare Solutions Inc., Osiris Therapeutics Inc., Otsuka America Pharmaceutical Inc., RemedyMD, Sanofi, Seattle Genetics, Sigma-Tao Pharmaceuticals, Soligenix Inc., Stem-Cyte, A Global Cord Blood Therapeutics Co., Stemsoft Software Inc., Swedish Orphan Biovitrum, Tarix Pharmaceuticals, Teva Neuroscience Inc., THERAKOS Inc., and Wellpoint Inc. The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
Authorship
Contribution: M.A.F.-V., T.W., S.J.L., M.H., S.R.S., and M.S. critically revised the research plan; M.A.F.-V., S.J.L., S.R.S., and M.S. drafted the manuscript; M.A.F.-V., T.W., S.J.L., M.H., M. Aljurf, M. Askar, M.B., L.B.-L., J.G., A.A.J., S.M., M.O., S.G.E.M., E.W.P., K.S., E.V.T., E.K.W., A.W., J.U., S.R.S., and M.S. analyzed and interpreted data and critically revised the manuscript; T.W., M.H., and J.U. performed statistics; and M.A.F.-V. and M.S. drafted the research plan.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcelo Fernandez-Viña, Department of Pathology, Stanford University School of Medicine, 3373 Hillview Ave, Palo Alto, CA 94304; e-mail: marcelof@stanford.edu.