Key Points
Molecular characterization of myeloma requires isolation of malignant plasma cells, which is currently hampered by the instability of CD138.
We identified CD319 and CD269 as robust replacements for CD138, facilitating molecular diagnostics in myeloma.
Abstract
Molecular characterization of malignant plasma cells is increasingly important for diagnostic and therapeutic stratification in multiple myeloma. However, the malignant plasma cells represent a relatively small subset of bone marrow cells, and need to be enriched prior to analysis. Currently, the cell surface marker CD138 (SDC1) is used for this enrichment, but has an important limitation in that its expression decreases rapidly after sampling. Seeking alternatives to CD138, we performed a computational screen for myeloma plasma cell markers and systematically evaluated 7 candidates. Our results conclusively show that the markers CD319 (SLAMF7/CS1) and CD269 (TNFRSF17/BCMA) are considerably more robust than CD138 and enable isolation of myeloma plasma cells under more diverse conditions, including the samples that have been delayed or frozen. Our results form the basis of improved procedures for characterizing cases of multiple myeloma in clinical practice.
Introduction
Multiple myeloma (MM) is characterized by the uncontrolled, clonal growth of plasma cells in the bone marrow. As our knowledge increases about the genetic basis of MM, and new therapeutic options are being developed, molecular characterization of MM plasma cells (MMPCs) is becoming increasingly important for subclassification, prognostication, and treatment stratification.1
Unlike disorders in which the malignant clone tends to dominate the bone marrow at diagnosis (eg, acute leukemia), MMPCs generally represent a relatively small subset of bone marrow cells. This means that the MMPCs need to be selected, or at least enriched, to ensure that results obtained with techniques, such as fluorescent in-situ hybridization, gene-expression profiling, copy-number profiling, and sequencing, are representative.2,3
In current practice, MMPCs are usually isolated using magnetic beads, or fluorescence-activated cell sorting (FACS), with antibodies toward CD138 (syndecan 1; SDC1), which is preferred because of its high plasma cell specificity,4,5 as compared with CD38 or CD56, for example.6 Nevertheless, the CD138 antigen has an important drawback in that it disappears rapidly from the cell surface when the analysis is delayed or when the sample is frozen.4,7 This phenomenon, attributed to rapid molecular turnover and shedding,8 limits the tolerable time between sampling and analysis, and reduces sensitivity.7 Although CD138 is applicable to freshly collected samples, robust MMPC markers would facilitate molecular MM diagnostics.
We undertook a computational screen based on large sets of gene expression microarray data to identify alternative MMPC cell surface markers. Seven candidate markers were evaluated. CD319 and CD269 proved to be robust replacements for CD138.
Study design
Microarray data analysis
Gene expression profiles of 1285 MM samples (CD138+ bone marrow cells) and 3164 examples of other hematologic malignancies (blood or bone marrow) were retrieved from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds; accession numbers GSE15695,9 GSE4581, GSE19784,10 GSE26760,2 GSE13159,11 GSE6891,12 GSE17855,13 and GSE1241714 ) and St. Jude Research15 (http://www.stjuderesearch.org/site/data/ALL3). All data were generated on Affymetrix U133Av2 microarrays and quantile-normalized to a normal distribution. Differentially expressed genes were identified using Smyth’s moderated t test.16 Affymetrix U133A expression profiles of 38 normal hematopoietic cell types were retrieved from the Differentiation Map (DMAP) (http://www.broadinstitute.org/dmap).17
Sample preparation and analysis
Both fresh and vital-frozen bone marrow samples were used (all from myeloma patients). Fresh samples were obtained from the Hematology Clinic, Skåne University Hospital, Lund; vital-frozen samples were obtained from the Swedish National Myeloma Biobank, Lund. The samples were stained as indicated using CD38-PE-Cy7(HB7), CD45-APC-Cy7(2D1), CD138-APC(MI15) (BD Biosciences), CD319-PE(162.1), CD307e-PE(509f6) (Biolegend), CD269-PE(goat anti-human; blocking) (R&D Systems), CD208-APC(31B), κ light chain-eFluor 450 (TB28-2), Λ light chain-PerCP-eFluor 710 (1-155-2) (eBioscience), GPRC5D-APC, ITGA8-APC(aa610-800) (LifeSpan BioSciences), and FKBP11(rabbit anti-human; LifeSpan BioSciences) conjugated with Alexa Fluor 488 (A20181; Life Technologies). FACS Aria II was used for analysis. DNA was purified using standard kits (QIAgen) and analyzed using Illumina HumanOmni5M-4 v1.0 copy-number arrays. The study was approved by the Ethical Review Board (Etikprövningsnämnden) at Lund University. The study was conducted in accordance with the Declaration of Helsinki.
Results and discussion
We retrieved 1285 gene expression profiles of CD138+ cells from MM bone marrow samples that had been previously collected in microarray studies.2,9,10 To enable identification of genes specifically expressed in MMPCs, we collected a control data set consisting of 3185 gene expression profiles from a wide range of hematologic disorders and cell types.11-15 Comparison of these 2 data sets identified a gene expression signature that is representative for a broad range of MMPCs (supplemental Table 1, available on the Blood Web site).
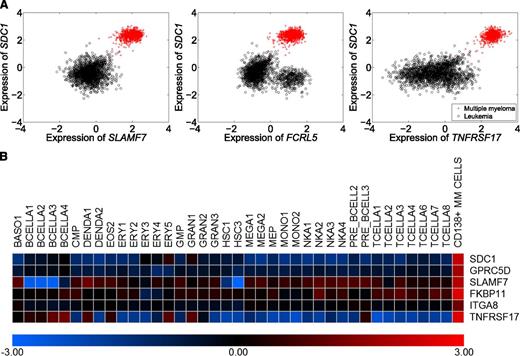
From these data, we identified TNFRSF17 (CD269/BCMA), SLAMF7 (CD319/CS1), GPRC5D, FKBP11, LAMP3 (CD208), ITGA8, and FCRL5 (CD307e) as encoding candidate MMPC markers (supplemental Table 1). These genes were selected because they were highly and specifically expressed in MMPCs, except FCRL5, which was also expressed in chronic lymphocytic leukemia (Figure 1A, data not shown). They were also selected because they encode likely cell-surface proteins because antibodies toward the respective gene products were available, and because they show higher expression levels in MMPCs than in any major normal hematopoietic cell type (Figure 1B).
Selection of candidate marker genes. To identify markers for further testing, we compared 1285 gene expression profiles of MMPCs with 3164 gene expression profiles of a wide range of hematologic disorders. Through this process, we identified large numbers of strongly differentially expressed genes (supplemental Table 1). Integrating the differential expression data with information about gene function, protein structure, and antibody availability, we selected 7 candidates for further testing: TNFRSF17 (CD269), SLAMF7 (CD319/CS1), GPRC5D, FKBP11, LAMP3 (CD208), ITGA8, and FCRL5 (CD307e). (A) Expression of 3 of our candidate marker genes, SLAMF7 (CD319), FCRL5 (CD307e), and TNFRSF17 (CD269) vs expression of SDC1 (CD138) in MM and leukemia (red, myeloma samples; black, control samples). All genes were more highly expressed in MMPCs and coexpressed with SDC1 (CD138). The results for GPRC5D, FKBP11, LAMP3, and ITGA8 are not shown, as their protein products could not be detected on MMPCs in the subsequent experiments. All genes were specifically expressed in MMPCs, except for FCRL5, which was also expressed in chronic lymphocytic leukemias (bottom-right cluster in middle panel). (B) Expression of our candidate genes in MMPCs and different normal hematopoietic cell types, represented here by gene expression profiles from the DMAP compendium. The selected genes were more highly expressed in MMPCs than in normal hematopoietic cells from different lineages. FCRL5 was not represented in DMAP.
Selection of candidate marker genes. To identify markers for further testing, we compared 1285 gene expression profiles of MMPCs with 3164 gene expression profiles of a wide range of hematologic disorders. Through this process, we identified large numbers of strongly differentially expressed genes (supplemental Table 1). Integrating the differential expression data with information about gene function, protein structure, and antibody availability, we selected 7 candidates for further testing: TNFRSF17 (CD269), SLAMF7 (CD319/CS1), GPRC5D, FKBP11, LAMP3 (CD208), ITGA8, and FCRL5 (CD307e). (A) Expression of 3 of our candidate marker genes, SLAMF7 (CD319), FCRL5 (CD307e), and TNFRSF17 (CD269) vs expression of SDC1 (CD138) in MM and leukemia (red, myeloma samples; black, control samples). All genes were more highly expressed in MMPCs and coexpressed with SDC1 (CD138). The results for GPRC5D, FKBP11, LAMP3, and ITGA8 are not shown, as their protein products could not be detected on MMPCs in the subsequent experiments. All genes were specifically expressed in MMPCs, except for FCRL5, which was also expressed in chronic lymphocytic leukemias (bottom-right cluster in middle panel). (B) Expression of our candidate genes in MMPCs and different normal hematopoietic cell types, represented here by gene expression profiles from the DMAP compendium. The selected genes were more highly expressed in MMPCs than in normal hematopoietic cells from different lineages. FCRL5 was not represented in DMAP.
CD269 has been previously identified as a regulator of B-cell development, and the receptor for tumor necrosis factor superfamily member 13b (TNFSF13B/BAFF).18 CD319 has been identified as a signaling molecule in plasma cells and certain lymphocyte subsets, and is the target of the anti-myeloma antibody elotuzumab.19,20 GPRC5D and ITGA8 have not been described as plasma cells cell markers. However, elevated GPRC5D transcript levels have been associated with poor-prognosis MM,21 and the ITGA8 transcript level decreases in MM cells treated with lenalidomide and pomalidomide.22 CD307e is expressed on plasma cells and has been suggested as a therapeutic target in MM.23-25 CD208 and FKBP11 have not been associated previously with plasma cells or MM. Importantly, none of the candidate markers has been previously evaluated with respect to its robustness under conditions that apply in diagnostic situations or biobanking.
To evaluate these markers, we quantified their protein levels by FACS in bone marrow aspirates from patients with MM that had been stored from 1 to 3 days at 8°C, mimicking the common clinical situation in which a sample cannot be immediately analyzed. We also analyzed protein levels in samples that had been vital-frozen, mimicking biobanking. Throughout, CD319, CD269, and CD307e defined discrete cell populations (and costained with CD138 in fresh samples), whereas CD208, GPRC5D, ITGA8, and FKBP11 could not be detected with the available antibodies (not shown).
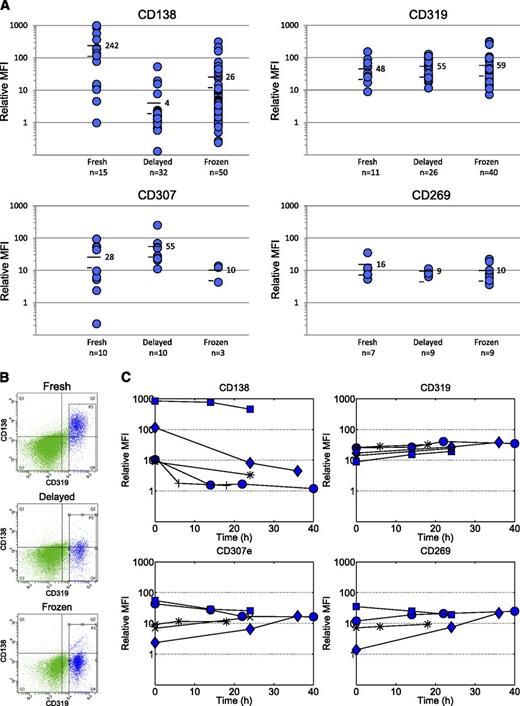
We tested larger numbers of fresh, delayed, or vital-frozen samples (as available) for CD319, CD269, or CD307e (plus CD138 and standard markers; see “Study design”). As anticipated, the CD138 level varied between samples and was considerably lower in delayed and frozen samples (average relative mean fluorescent intensity (MFI) 4 and 26, respectively) than in fresh samples (average relative MFI 242) (Figure 2A). Many samples showed a relative MFI close to 1, meaning CD138 could not discriminate MMPCs from other cells.
Evaluation of MMPC markers with respect to robustness. Three candidate markers (CD319, CD307e, and CD269) allowed identification of MMPCs, whereas 4 markers (CD208, GPRC5D, ITGA8, and FKBP11) could not be detected. (A) Protein levels on MMPCs expressed as MFI relative to other cells in samples that had been stored for 6 to 24 hours at 8°C or vital-frozen prior to analysis. As expected, CD138 could be detected at variable levels in fresh samples, but showed considerably lower expression in stored/delayed and frozen samples. By contrast, CD319 and CD269 showed remarkably stable expression (relative MFI approximately 10-100 and approximately 10, respectively) and enabled the identification of a discrete, marker-positive population. Cells isolated from these populations by FACS were confirmed to be MMPCs by costaining with CD138 in fresh samples, clonal excess analysis, light microscopy, and DNA copy number microarray (supplemental Figure 1). CD307e showed more varied expression. (B) Representative example showing that both CD138 and CD319 are present on MMPCs in fresh samples. However, when the same sample was analyzed the following day or after vital-freezing, CD138 was lost, but CD319 was retained. The figure also illustrates how MMPCs (blue events) were discriminated from other cells (green events). (C) Time-series data, in which the same samples were stained/analyzed at varying time points, were between 0 and 40 hours of storage at 8°C.
Evaluation of MMPC markers with respect to robustness. Three candidate markers (CD319, CD307e, and CD269) allowed identification of MMPCs, whereas 4 markers (CD208, GPRC5D, ITGA8, and FKBP11) could not be detected. (A) Protein levels on MMPCs expressed as MFI relative to other cells in samples that had been stored for 6 to 24 hours at 8°C or vital-frozen prior to analysis. As expected, CD138 could be detected at variable levels in fresh samples, but showed considerably lower expression in stored/delayed and frozen samples. By contrast, CD319 and CD269 showed remarkably stable expression (relative MFI approximately 10-100 and approximately 10, respectively) and enabled the identification of a discrete, marker-positive population. Cells isolated from these populations by FACS were confirmed to be MMPCs by costaining with CD138 in fresh samples, clonal excess analysis, light microscopy, and DNA copy number microarray (supplemental Figure 1). CD307e showed more varied expression. (B) Representative example showing that both CD138 and CD319 are present on MMPCs in fresh samples. However, when the same sample was analyzed the following day or after vital-freezing, CD138 was lost, but CD319 was retained. The figure also illustrates how MMPCs (blue events) were discriminated from other cells (green events). (C) Time-series data, in which the same samples were stained/analyzed at varying time points, were between 0 and 40 hours of storage at 8°C.
By contrast, with CD319 and CD269, we consistently identified marker-positive cell populations in both delayed and frozen samples, including samples where CD138-positive cells could not be detected (Figure 2A,B and supplemental Figure 1). To confirm that the identified cell populations were MMPCs, we isolated CD319-positive and CD269-positive cells in selected samples by FACS. Marker-positive cells showed MMPC morphology, clonal excess, and myeloma-associated gains or losses of chromosomal material by DNA copy number microarray (supplemental Figures 2 and 3). CD307e was present at variable levels, increased during storage, and decreased during freezing (Figure 2A). All 3 markers were expressed on plasma cells from healthy donors (supplemental Figure 4).
To investigate stability over time, we stained samples from 5 cases for CD138, CD269, CD307e, and CD319 at time points between 0 and 40 hours of storage at 8°C. Consistent with Figure 2A, CD138 decreased in 4 of 5 cases, whereas CD319 persisted in all cases, and CD269 persisted in almost all but 1 case, where the initial level was unexpectedly low (Figure 2C).
Characterization of MMPCs is rapidly becoming important for subclassification and clinical decision making in MM, but requires the enrichment of MMPCs to yield representative results. The fact that CD138 is unstable complicates enrichment, limits the acceptable time to analysis, and reduces sensitivity.
We show that CD319 and CD269 enable isolation of MMPCs under substantially more diverse conditions than the current standard marker CD138, including within delayed samples, which are common in practice, and vital-frozen samples, enabling studies of banked, unsorted samples. CD319 and CD269 have been reported to be expressed on MMPCs, but their ability to isolate MMPCs robustly has not been previously demonstrated.
Taken together, our results identify CD319 and CD269 as robust replacements for CD138, and point the way to improved procedures (eg, novel magnetic bead-based purification protocols) for analyzing MMPCs in clinical practice.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research grants from the Swedish Foundation for Strategic Reseearch (KF10-0009 and ICA08-0057), the Marianne and Marcus Wallenberg Foundation (2010.0112), the Knut and Alice Wallenberg Foundation (2012.0193), the Royal Swedish Academy of Science, Avtal om Läkares Forskningstid grants to the University and Regional Laboratories (Labmedicin Skåne), the Siv-Inger and Per-Erik Andersson Foundation, the Medical Faculty at Lund University, and the Swedish Society of Medicine.
Authorship
Contribution: B.N., M.H., U.G., and J.A. designed the research; I.F., J.A., E.J., M.A., and M.K.C. performed experiments and/or data analysis; M.H. and I.T. provided samples; and B.N., M.H., U.G., and I.F. wrote the paper. All authors contributed to data interpretation and to the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Björn Nilsson, Hematology and Transfusion Medicine, Department of Laboratory Medicine, BMC B13, Lund, 22184 Sweden; e-mail: bjorn.nilsson@med.lu.se.
References
Author notes
I.F. and J.A. contributed equally to this study.
M.H. and B.N. contributed equally to this study.