Key Points
HDACi-mediated differentiation therapy is a potent and molecularly rational treatment strategy in t(8;21) AML.
Abstract
Epigenetic modifying enzymes such as histone deacetylases (HDACs), p300, and PRMT1 are recruited by AML1/ETO, the pathogenic protein for t(8;21) acute myeloid leukemia (AML), providing a strong molecular rationale for targeting these enzymes to treat this disease. Although early phase clinical assessment indicated that treatment with HDAC inhibitors (HDACis) may be effective in t(8;21) AML patients, rigorous preclinical studies to identify the molecular and biological events that may determine therapeutic responses have not been performed. Using an AML mouse model driven by expression of AML1/ETO9a (A/E9a), we demonstrated that treatment of mice bearing t(8;21) AML with the HDACi panobinostat caused a robust antileukemic response that did not require functional p53 nor activation of conventional apoptotic pathways. Panobinostat triggered terminal myeloid differentiation via proteasomal degradation of A/E9a. Importantly, conditional A/E9a deletion phenocopied the effects of panobinostat and other HDACis, indicating that destabilization of A/E9a is critical for the antileukemic activity of these agents.
Introduction
Modulation of chromatin through histone modification is essential in regulating fundamental biological processes such as gene transcription and DNA repair.1 The biological consequences are determined by the combinatorial pattern of the histone modifications, which is regulated by specific enzymes that are classified into “writers,” “erasers,” and “readers.” Writers, such as histone acetyltransferases (HATs) or histone methyltransferases (HMTs), add distinct chemical modifications to histones; whereas erasers, including histone deacetylases (HDACs) and histone demethylases, remove these modifications. Lastly, epigenetic readers specifically recognize modified histones and recruit various effector molecules involved in transcriptional and chromatin regulation.2 Lysine acetylation, one of the major regulatory histone modifications, is controlled by the opposing activities of HATs and HDACs.3,4 In addition to modifying gene transcription via histone acetylation, HATs/HDACs also target nonhistone proteins affecting their stability, localization, and function.5,6 In mammals, 18 HDACs have been identified, which can be divided into 4 classes based on structure and cellular localization.4 The majority of these HDACs (ie, class I, II, and IV) depend on Zn2+ for their activity, whereas only class III HDACs (referred to as sirtuins) require NAD+.
Although cancer historically has been viewed as a disease originating from genetic alterations, recent findings have implicated epigenetic aberrations in the initiation and progression of human cancer.7 For example, expression of the tumor-suppressor genes CDKN2A, APC, and MLH1 is commonly silenced through promoter hypermethylation; whereas in hematologic cancers histone-modifying enzymes such as HDACs and HMTs are aberrantly localized to the genome through recruitment via oncogenic fusion proteins.7,8 The recent discovery of recurrent mutations in genes encoding the DNA methylation regulators DNMT3A and TET2, the HMT EZH2, and the HATs CREBBP and EP300 in human tumors further underpin the importance of epigenetic aberrations in tumorigenesis. Unlike genetic mutations, these are potentially reversible, implicating approaches to target epigenetic writers, erasers, and readers as promising and feasible strategies for cancer therapy.2,9
Numerous HDAC inhibitors (HDACis) have been developed, the majority of which lack isoform selectivity and broadly inhibit various Zn2+-dependent HDACs.10,11 HDACis demonstrate single-agent clinical activity against various hematologic malignancies, including different T-cell lymphomas and acute myeloid leukemia (AML),12-18 which has led to the US Food and Drug Administration (FDA) approval of vorinostat (targeting class I, II, and IV HDACs) as well as the more selective HDACi romidepsin (class I HDAC specific) for treatment of cutaneous T-cell lymphoma. The exact mechanism(s) responsible for the observed HDACi-mediated antitumor activity is still largely unclear and may depend on tumor type and the tumor genetic background.
The AML1-ETO fusion protein produced as a result of the t(8;21) chromosomal translocation is pathogenic for AML in collaboration with secondary mutagenic hits to genes such as FLT3, c-KIT, N-RAS, and K-RAS.19 AML1-ETO recruits a vast array of transcription factors and epigenetic regulatory proteins (eg, C/EBPα, GATA1, E proteins, HDACs, p300, PRMT1, and SON) and complexes (eg, N-CoR and AETFC)20-29 that putatively play important roles in the onset and/or progression of t(8;21) AML. Several studies have proposed the therapeutic potential of HDACis in t(8;21) AML,30-33 but some controversy exists regarding the mechanism by which HDACis mediate their effect. Treatment of A/E-positive Kasumi-1 cells with HDACis in vitro leads to tumor cell differentiation, which in some instances is accompanied by apoptosis.32,33 In contrast, the single study so far that has tested HDACis in vivo for t(8;21) AML indicated that cellular differentiation was not essential for the therapeutic effect observed. That study suggested that HDACi efficacy relied on the extrinsic apoptotic pathway mediated primarily by TNF-related apoptosis-inducing ligand (TRAIL) and its cognate receptor (DR5), which are both upregulated by HDACis in a tumor cell–selective manner.31 A potential confounding issue with most published studies testing the applicability of HDACis in t(8;21) AML is the use of valproic acid (VPA) as the HDACi. VPA is at best a weak HDACi34 resulting in its use at millimolar concentrations to induce histone hyperacetylation, raising concerns about potential off-target effects.
Herein we used a recently developed tractable mouse model of t(8;21) AML35 to identify the molecular and biological events that underpin the therapeutic effects of HDACi. We focused on panobinostat, which has strong inhibitory activity at low nanomolar concentrations against class I, II, and IV HDACs.34,36 In this transplantable model, AML develops as a result of the coexpression of AML1/ETO9a (A/E9a)37 and oncogenic Nras (NrasG12D). In A/E9a;NrasG12D-driven leukemia, HDAC inhibition triggered proteasomal degradation of AE/9a leading to terminal differentiation, subsequent decrease in tumor burden, and therapeutic efficacy. To our knowledge, this is the first definitive evidence that HDACi-mediated differentiation of t(8;21) AML cells results in a clear therapeutic benefit. Moreover, we have identified the biological and molecular processes that underpin the therapeutic effects of HDACis in this setting.
Materials and methods
Experimental animals and materials
C57BL/6 and C57BL/6.SJL-Ptprca mice were purchased from the Walter and Eliza Hall Institute of Medical Research and Animal Resources Centre, respectively. Nzeg-enhanced GFP (green fluorescent protein) (eGFP) mice were obtained from Prof Klaus Matthaei (Australian National University). All mice were used in accordance with the institutional guidelines of the Peter MacCallum Cancer Centre. Panobinostat was provided by Novartis and prepared as a 2-mg/mL solution in 5% dextrose/dH2O (D5W). Cytarabine was obtained from the Peter MacCallum Cancer Centre and further diluted in phosphate-buffered saline to a 12.5-mg/mL solution.
Fetal liver cell isolation, retroviral transduction, and transplantation
E13.5-15 fetal liver cells were cultured in the presence of interleukin (IL) 3, IL-6, and stem cell factor. For production of retrovirus-containing supernatant, Phoenix packaging cells were transfected with MSCV-AML1/ETO9a-IRES-GFP (A/E9a), MSCV-luciferase-IRES-NrasG12D, MSCV-MLL-AF9-IRES-VENUS (M/A), MSCV-MLL-ENL-IRES-GFP (M/E), MSCV-Bcl-2-IRES-mCherry, TREtight-dsRed-IRES-AML1/ETO9a, or MSCV-NrasG12D-IRES-tTA expression constructs. Supernatants were mixed in a 1:1 ratio and used to transduce fetal liver cells in 6-well plates precoated with RetroNectin (TaKaRa Bio). Two days after the last viral hit, 1 × 106 total cells per recipient were injected intravenously into lethally irradiated C57BL/6 mice.
Monitoring leukemia
Whole-body bioluminescent imaging (BLI) was assessed with an IVIS100 imaging system (Caliper LifeSciences). Mice were injected intraperitoneally with 50 mg/kg D-luciferin (Caliper LifeSciences), anesthetized with isoflurane, and imaged for 2 minutes after a 15-minute incubation following injection. Blood (∼30 µL) was obtained from the retro-orbital sinus. White cell counts were measured using an Advia 120 automated hematology analyzer (Bayer Diagnostics), and percentage of GFP-positive or dsRed-positive cells was analyzed by flow cytometry. At the terminal disease stage, mice were euthanized, and leukemia cells were isolated from bone marrow (femur) and spleen. Single-cell suspensions were prepared, and cells were cryopreserved in fetal calf serum (FCS)/10% dimethylsulfoxide (DMSO).
In vivo experiments
Leukemia cells (1 × 106) were transplanted into nonirradiated (M/E;NrasG12D and M/A;NrasG12D leukemias) or sublethally irradiated (A/E9a;NrasG12D leukemias) mice by intravenous injection. Treatment was initiated once leukemia was clearly established as demonstrated by visible bioluminescence and/or when there was 5% to 20% GFP-positive cells in peripheral blood. Mice were treated daily with panobinostat (25 mg/kg, 5 consecutive days per week intraperitoneal injection for 1 week followed by 15 mg/kg for 3 to 4 weeks) or vorinostat (200 mg/kg, 7 consecutive days per week intraperitoneally for 1 week followed by 150 mg/kg for 3 to 4 weeks). This dosing schedule of panobinostat was well tolerated by recipient mice (supplemental Figure 1A; see the Blood Web site). Control mice received the equivalent volume of vehicle. Mortality events from advancing leukemia were recorded for the analysis of therapeutic efficacy.
For short-term drug response studies, mice received 25 mg/kg panobinostat, 100 mg/kg cytarabine, or the equivalent volume of vehicle by intraperitoneal injection once daily for the indicated period prior to harvesting of spleen and bone marrow.
For inducible knockdown studies, mice received 2 mg/mL doxycycline via drinking water (2% saccharose) and food 2 weeks after reconstitution of mice with transduced Nzeg-eGFP fetal liver cells.
Flow cytometry
Cell suspensions were incubated in red cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) and washed twice in fluorescence-activated cell sorter (FACS) staining buffer (phosphate-buffered saline supplemented with 1% FCS and 0.02% NaN3). Cells were preincubated with anti-CD16/CD32 (2.4G2) and stained on ice with antibodies specific for c-Kit (CD117) and Mac-1 (CD11b) (both BD Biosciences) in FACS staining buffer for 30 minutes. Data were collected on a FACSCanto II flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Cell culture
Kasumi cells were cultured in RPMI 1640 + 10% FCS; A/E9a cells were cultured in a high-glucose version of Dulbecco’s modified Eagle medium supplemented with 10% FCS; and primary human t(8;21) cells were cultured at a starting density of 7.5 × 105 cells per mL in serum-free media consisting of Iscove modified Dulbecco medium supplemented with 20% BIT 9500 Serum Substitute (Stem Cell Technologies), 100 ng/mL stem cell factor, 100 ng/mL fms-like tyrosine kinase 3 ligand, 100 ng/mL thrombopoietin, and 20 ng/mL IL-6 (all Peprotech). Cells were treated with panobinostat for 6 or 24 hours at the indicated concentration.
Western blot analysis and immunoprecipitation
Cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% nonfat milk in Tris-buffered saline/0.1% Tween-20 at room temperature and incubated overnight with antibodies against AML1 (4336; Cell Signaling Technology), HDAC1 (5356; Cell Signaling Technology), p16 (M-156; Santa Cruz Biotechnology), p21 (F-5; Santa Cruz Biotechnology), phosphorylated retinoblastoma (RB) (554136; BD Biosciences), GFP (A6455; Invitrogen), histone H3 (AB1791; Abcam), acetyl-histone H3 (06-599; Millipore), heat shock protein 90 (Hsp90) (SPA830; Stressgen), glyceraldehyde-3-phosphate dehydrogenase (AB9484; Abcam), or β-actin (A2228; Sigma-Aldrich) at 4°C. Membranes were developed using appropriate horseradish peroxidase–coupled secondary antibodies (Dako) and enhanced chemiluminescence (GE Healthcare).
For immunoprecipitations, cell lysates were precleared by incubation with protein A or G sepharose beads (GE Healthcare) for 30 minutes, washed in lysis buffer, incubated with anti-AML1 or anti-Hsp90 antibodies for 30 minutes, and then cross-linked to protein A or G sepharose beads at 4°C overnight. Beads were washed 5 times with lysis buffer before addition of 5× denaturing loading buffer, heating at 95°C, and analysis by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Quantitative reverse-transcription polymerase chain reaction
RNA was isolated using a commercially available kit (Qiagen). Synthesis of complementary DNA was performed following standard protocols using MMLV Reverse Transcriptase (Promega). Quantitative reverse-transcription PCR was performed using the SYBR Green (Applied Biosystems) method in a 384-well format using the ABI Prism7900HT (Applied Biosystems). For quantification, the CT values were obtained and normalized to the CT values of the HPRT gene. Fold changes in expression were calculated by the 2–ΔΔCT method.
Histology
Mice femora were fixed in 10% neutral buffered formalin before decalcification using formic acid and embedding in paraffin. Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining was performed using the Apoptag Peroxidase In Situ Apoptosis Detection Kit (Chemicon) kit. Sections were counterstained using hematoxylin, and images were recorded with a Zeiss microscope and a ×20 lens. For cytospins, bone marrow cells were stained with Romanowski stain solution.
Statistical analysis
Kaplan-Meier survival curves were created, and survival of mice (using a log-rank test) was statistically analyzed with GraphPad Prism software (GraphPad software). All other statistical analyses (using a 2-tailed unpaired Student t test) were also performed with this software.
Results
A/E9a;NrasG12D-driven leukemia is susceptible to the HDACi panobinostat
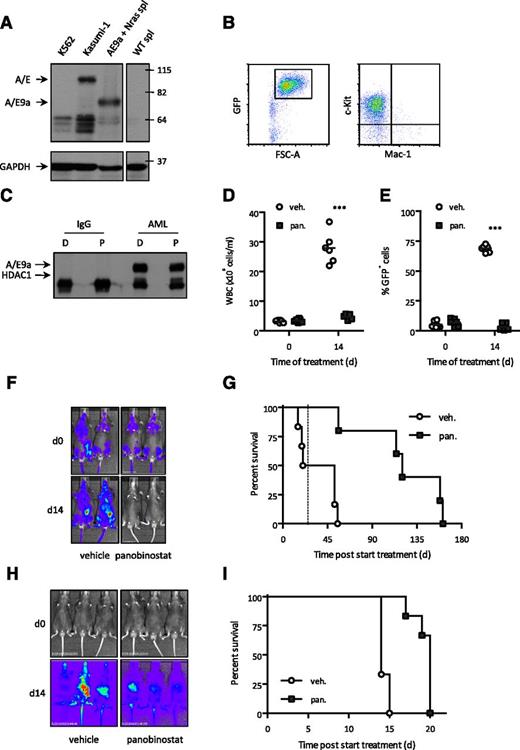
Recipient mice transplanted with fetal liver cells transduced with retroviral vectors encoding A/E9a (coexpressing GFP) and NrasG12D (coexpressing firefly luciferase) developed AML. Leukemic cells expressed A/E9a (Figure 1A), were GFP positive (Figure 1B), and expressed c-Kit (CD117), but not Mac-1 (CD11b) or other lineage markers (Figure 1B and data not shown). Consistent with the previously identified interaction between A/E and HDACs,20,21,38-40 immunoprecipitation assays confirmed the interaction between HDAC1 and A/E9a in primary A/E9a;NrasG12D leukemic cells (Figure 1C). Similar experiments were performed to determine if HDAC2 and HDAC3 also associated with A/E9a, and although HDAC2 immunoprecipitated with A/E9a, no physical interaction was detected between HDAC3 and A/E9a (data not shown). Treatment with panobinostat had no acute effect on the interaction of HDACs with A/E9a (Figure 1C and data not shown).
The HDACi panobinostat demonstrates therapeutic efficacy in a mouse model of A/E9a;NrasG12D-driven but not M/E;NrasG12D-driven AML. (A) Western blot analysis of whole-cell lysates prepared from cell lines K562 and Kasumi-1 and spleen cells (spl) isolated from a WT and an A/E9a;NrasG12D leukemia recipient mouse using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control (bottom panel). (B) Flow cytometry analysis of c-Kit and Mac-1 expression in GFP-positive spleen cells isolated from an A/E9a;NrasG12D leukemia recipient mouse. A representative flow cytometry plot is shown. (C) Immunoprecipitation/western blot analysis of the interaction between A/E9a and HDAC1 in A/E9a;NrasG12D leukemic cells treated with DMSO (D) or 16 nM panobinostat (P) for 6 hours. A control mouse immunoglobulin G (IgG) and antibody to AML1 (AML) were used for immunoprecipitation; antibodies to HDAC1 and AML1 were used for western blotting. The results shown are representative of 3 independent experiments. (D-F) Total white blood cells (WBC), flow cytometry analysis of leukemic cells in peripheral blood, and BLI of C57BL/6 mice bearing A/E9a;NrasG12D tumors treated with panobinostat or vehicle using the standard treatment regimen. In panels D and E, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (G) Kaplan-Meier survival curves of treated A/E9a;NrasG12D leukemia recipient mice following initiation of therapy (n = 6 for vehicle, n = 5 for panobinostat; median survival benefit 84 days, P = .0006). Dotted line indicates final day of treatment. (H) BLI of M/E;NrasG12D leukemia recipient mice treated with panobinostat or vehicle using the standard treatment regimen. (I) Kaplan-Meier survival curves of treated M/E;NrasG12D leukemia recipient mice following initiation of therapy (n = 6 for vehicle, n = 6 for panobinostat; median survival benefit 6 days, P = .0015).
The HDACi panobinostat demonstrates therapeutic efficacy in a mouse model of A/E9a;NrasG12D-driven but not M/E;NrasG12D-driven AML. (A) Western blot analysis of whole-cell lysates prepared from cell lines K562 and Kasumi-1 and spleen cells (spl) isolated from a WT and an A/E9a;NrasG12D leukemia recipient mouse using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control (bottom panel). (B) Flow cytometry analysis of c-Kit and Mac-1 expression in GFP-positive spleen cells isolated from an A/E9a;NrasG12D leukemia recipient mouse. A representative flow cytometry plot is shown. (C) Immunoprecipitation/western blot analysis of the interaction between A/E9a and HDAC1 in A/E9a;NrasG12D leukemic cells treated with DMSO (D) or 16 nM panobinostat (P) for 6 hours. A control mouse immunoglobulin G (IgG) and antibody to AML1 (AML) were used for immunoprecipitation; antibodies to HDAC1 and AML1 were used for western blotting. The results shown are representative of 3 independent experiments. (D-F) Total white blood cells (WBC), flow cytometry analysis of leukemic cells in peripheral blood, and BLI of C57BL/6 mice bearing A/E9a;NrasG12D tumors treated with panobinostat or vehicle using the standard treatment regimen. In panels D and E, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (G) Kaplan-Meier survival curves of treated A/E9a;NrasG12D leukemia recipient mice following initiation of therapy (n = 6 for vehicle, n = 5 for panobinostat; median survival benefit 84 days, P = .0006). Dotted line indicates final day of treatment. (H) BLI of M/E;NrasG12D leukemia recipient mice treated with panobinostat or vehicle using the standard treatment regimen. (I) Kaplan-Meier survival curves of treated M/E;NrasG12D leukemia recipient mice following initiation of therapy (n = 6 for vehicle, n = 6 for panobinostat; median survival benefit 6 days, P = .0015).
C57BL/6 mice transplanted with leukemia cells from a single A/E9a;NrasG12D leukemia mouse demonstrated a clear reduction in tumor burden following panobinostat treatment (Figure 1D-F) resulting in a pronounced survival benefit (Figure 1G). Similar therapeutic responses with panobinostat were observed in C57BL/6 mice transplanted with cells from a different A/E9a;NrasG12D leukemia mouse (supplemental Figure 1B). Treatment of wild-type (WT) C57BL/6 mice with panobinostat using the same treatment regimen as used for the treatment of tumor-bearing mice resulted in a slight decrease in peripheral white blood counts and little or no weight loss or other signs of toxicity (Salmon and Johnstone, unpublished observations). Moreover, vorinostat, an FDA-approved HDACi, also mediated a reduction in tumor burden and prolonged survival of A/E9a;NrasG12D tumor-bearing mice (supplemental Figure 1C-D). We additionally determined if chronic exposure to panobinostat in vivo altered the phenotype of A/E9a;NrasG12D tumors or induced acquired resistance to the compound. A/E9a;NrasG12D tumors harvested from tumor-bearing mice treated with panobinostat for 4 cycles were assessed by flow cytometry for GFP expression and expression for c-Kit and Mac-1. As shown in supplemental Figure 1E, cells chronically exposed to panobinostat retained their immature AML phenotype. Moreover, mice transplanted with GFP-positive A/E9a;NrasG12D tumors harvested from mice that had received 8 cycles of panobinostat treatment and subsequently treated with panobinostat showed a robust therapeutic response (supplemental Figure 1F), similar to mice bearing A/E9a;NrasG12D tumors not previously exposed to panobinostat.
To determine whether panobinostat was also effective in other AML subtypes, we generated leukemias in mice coexpressing the mixed lineage leukemia (MLL) fusion proteins MLL/ENL (M/E) or MLL/AF9 (M/A) and NrasG12D. Recipient mice were transplanted with leukemic cells, and mice bearing secondary transplanted tumors were treated with panobinostat resulting in a marginal effect on leukemia progression and survival (Figure 1H-I and supplemental Figure 2A-C).
Response to panobinostat is independent of a functional p53 pathway
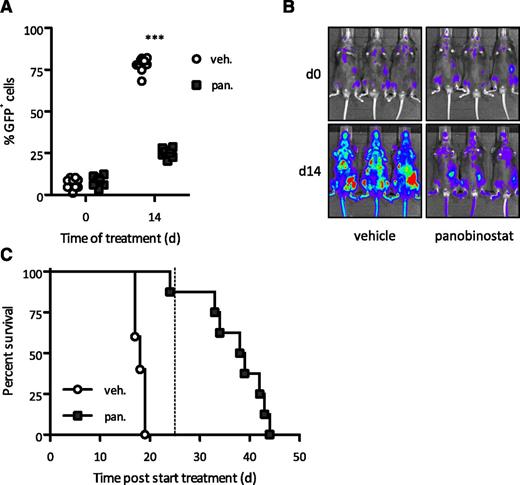
Standard treatment of AML patients currently consists of cytarabine in combination with an anthracycline such as doxorubicin.41 Many standard chemotherapeutics rely on an intact p53 pathway for an optimal therapeutic response,42 and we have shown that p53-deficient A/E9a;NrasG12D leukemias were refractory to standard chemotherapy35 (supplemental Figure 3A). We previously showed that the apoptotic and therapeutic effects of HDACi's in a B-cell lymphoma model occurred in the absence of WT p53,43,44 and we wished to investigate the response of p53-deficient A/E9a;NrasG12D leukemias to panobinostat. Panobinostat reduced the leukemic burden and significantly prolonged the survival of mice bearing highly aggressive A/E9a;NrasG12D/p53−/− leukemias (Figure 2A-C and supplemental Figure 3B). These results suggest that HDACis may provide a therapeutic option in p53-deficient AML that is commonly resistant to conventional induction therapy.45
Response to panobinostat is independent of a functional p53 pathway. (A-B) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/p53−/− leukemia recipient mice treated with panobinostat or vehicle using the standard treatment regimen. In panel A, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (C) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/p53−/− leukemia recipient mice following initiation of therapy (n = 10 for vehicle, n = 8 for panobinostat; median survival benefit 21 days, P < .0001). Dotted line indicates final day of treatment.
Response to panobinostat is independent of a functional p53 pathway. (A-B) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/p53−/− leukemia recipient mice treated with panobinostat or vehicle using the standard treatment regimen. In panel A, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (C) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/p53−/− leukemia recipient mice following initiation of therapy (n = 10 for vehicle, n = 8 for panobinostat; median survival benefit 21 days, P < .0001). Dotted line indicates final day of treatment.
Induction of apoptosis is dispensable for a therapeutic response to panobinostat
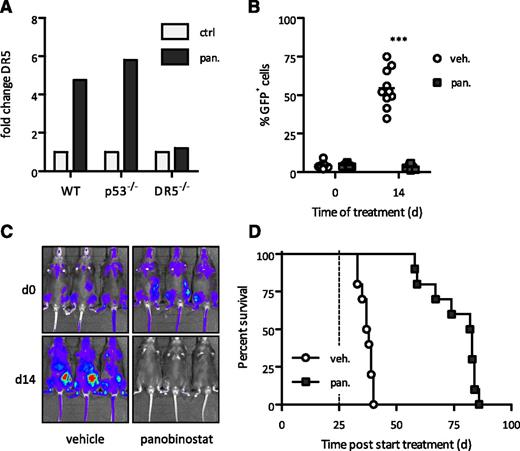
Based on previous studies showing a direct link between HDACi-induced tumor cell apoptosis and preclinical efficacy,31,38,43,44,46,47 we generated A/E9a;NrasG12D leukemias with defined genetic alterations in both the extrinsic (knockout of either TRAIL or its receptor DR5) and the intrinsic (overexpressing Bcl-2, supplemental Figure 4A) apoptotic pathways. Although Dr5 expression increased upon treatment with panobinostat (Figure 3A), A/E9a;NrasG12D leukemias with deletion of Dr5 (Figure 3B-C) or Trail (supplemental Figure 5A-B) retained profound sensitivity to HDACi treatment. Importantly, the potent antileukemic activity of panobinostat was comparable in independently derived DR5−/− or TRAIL−/− A/E9a;NrasG12D leukemias (Figure 3D and supplemental Figures 5C-D and 6). These data provide definitive evidence that a functional TRAIL pathway is not required for HDACi to mediate a robust therapeutic effect against t(8;21) AML.
Extrinsic apoptotic pathway is dispensable for therapeutic response of panobinostat. (A) Quantitative real-time PCR of Dr5 messenger RNA (mRNA) levels in spleen cells (>80% GFP-positive cells) isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W) for 4 hours. Mean value of 2 individual samples is shown. (B-C) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/DR5−/− leukemia recipient mice treated with panobinostat or vehicle using the standard therapy regimen. In panel B, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (D) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/DR5−/− leukemia recipient mice following initiation of therapy (n = 10 for vehicle, n = 10 for panobinostat; median survival benefit 45 days, P < .0001). Dotted line represents final day of treatment.
Extrinsic apoptotic pathway is dispensable for therapeutic response of panobinostat. (A) Quantitative real-time PCR of Dr5 messenger RNA (mRNA) levels in spleen cells (>80% GFP-positive cells) isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W) for 4 hours. Mean value of 2 individual samples is shown. (B-C) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/DR5−/− leukemia recipient mice treated with panobinostat or vehicle using the standard therapy regimen. In panel B, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (D) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/DR5−/− leukemia recipient mice following initiation of therapy (n = 10 for vehicle, n = 10 for panobinostat; median survival benefit 45 days, P < .0001). Dotted line represents final day of treatment.
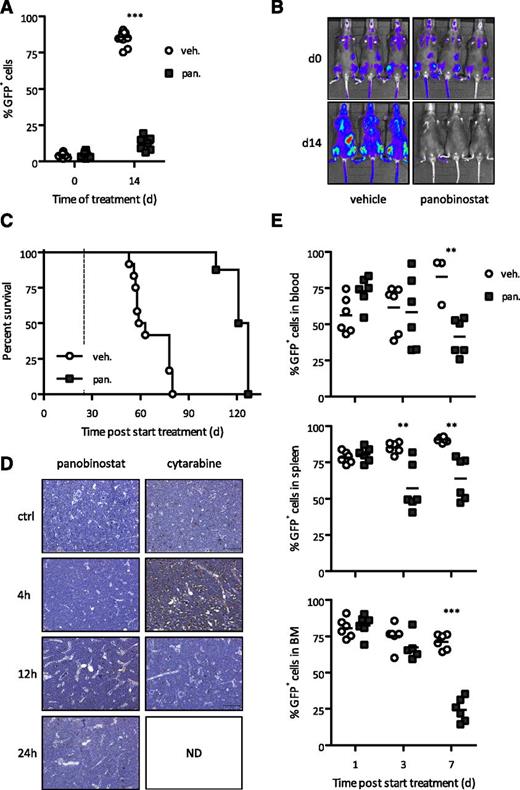
Overexpression of Bcl-2 (supplemental Figure 4A) also had no effect on the sensitivity of A/E9a;NrasG12D leukemias to panobinostat in vivo (Figure 4A-B). Mice bearing these leukemias treated with panobinostat clearly had a survival benefit over vehicle-treated mice (Figure 4C and supplemental Figure 4B). These data imply that tumor cell apoptosis was not the primary biological response of A/E9a;NrasG12D leukemias to panobinostat. Consistent with this observation, few or no TUNEL-positive tumor cells were detected in leukemia-bearing mice following treatment with panobinostat (Figure 4D and supplemental Figure 7). In contrast, treatment with cytarabine led to a substantial increase in TUNEL-positive tumor cells (Figure 4D and supplemental Figure 7). Despite their failure to undergo apoptosis in response to panobinostat, A/E9a;NrasG12D leukemias were reduced in spleen, bone marrow, and peripheral blood within 3 to 7 days posttreatment (Figure 4E). Hence, although A/E9a;NrasG12D leukemias can undergo apoptosis in response to certain anticancer agents, our data strongly implicate a nonapoptotic primary effector mechanism for panobinostat.
Intrinsic apoptotic pathway is dispensable for therapeutic response to panobinostat. (A-B) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/Bcl-2 leukemia recipient mice treated with panobinostat or vehicle using the standard therapy regimen. In panel A, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (C) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/Bcl-2 leukemia recipient mice following initiation of therapy (n = 12 for vehicle, n = 8 for panobinostat; median survival benefit 63 days, P < .0001). Dotted line represents final day of treatment. (D) Analysis of apoptotic cells via TUNEL staining. Staining was performed on bone marrow isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or cytarabine (100 mg/kg) for the indicated time. Sections are representative of 3 (panobinostat) or 2 (cytarabine) mice per time point. Dark brown cells indicate TUNEL-positive cells. (E) Flow cytometry analysis of GFP-positive leukemic cells in indicated tissue isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W, 250 µL) for 3 days. Data are combined from 2 individual experiments. Each data point represents an individual mouse, and horizontal bars represent mean value. **P = .004 (blood); **P = .0022 and P = .0012, respectively (spleen); ***P < .0001 (bone marrow).
Intrinsic apoptotic pathway is dispensable for therapeutic response to panobinostat. (A-B) Flow cytometry analysis of leukemic cells in peripheral blood and BLI of A/E9a;NrasG12D/Bcl-2 leukemia recipient mice treated with panobinostat or vehicle using the standard therapy regimen. In panel A, each data point represents an individual mouse, and horizontal bars represent mean value. ***P < .0001. (C) Kaplan-Meier survival curves of treated A/E9a;NrasG12D/Bcl-2 leukemia recipient mice following initiation of therapy (n = 12 for vehicle, n = 8 for panobinostat; median survival benefit 63 days, P < .0001). Dotted line represents final day of treatment. (D) Analysis of apoptotic cells via TUNEL staining. Staining was performed on bone marrow isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or cytarabine (100 mg/kg) for the indicated time. Sections are representative of 3 (panobinostat) or 2 (cytarabine) mice per time point. Dark brown cells indicate TUNEL-positive cells. (E) Flow cytometry analysis of GFP-positive leukemic cells in indicated tissue isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W, 250 µL) for 3 days. Data are combined from 2 individual experiments. Each data point represents an individual mouse, and horizontal bars represent mean value. **P = .004 (blood); **P = .0022 and P = .0012, respectively (spleen); ***P < .0001 (bone marrow).
Panobinostat induces differentiation of A/E9a;NrasG12D AML
Treatment of primary A/E9a;NrasG12D leukemias with panobinostat in vitro resulted in a reduction in the number of cells in S phase (Figure 5A) and biochemical changes characteristic with a block in cell cycle at the G1/S checkpoint including increased expression of p21WAF1/CIP1 and p16INK4A and hypophosphorylation of pRb (Figure 5B). These changes in cell proliferation were concomitant with a decrease in expression of c-Kit following treatment with panobinostat for 24 or 48 hours (Figure 5C). No change in c-Kit expression was seen following treatment of A/E9a;NrasG12D cells with etoposide (Figure 5C) demonstrating the specific effect of panobinostat. Concomitant with panobinostat-mediated induction of tumor cell cycle arrest and decreased expression of c-Kit, an increase in mRNA for the promyeloid differentiation transcription factors PU-1, GATA-2, SCL, and C/EBP alpha was observed (Figure 5D).
Panobinostat treatment of A/E9a;NrasG12Dleukemic cells triggers differentiation. (A) Cell cycle analysis of A/E9a;NrasG12D leukemic cells treated in vitro with vehicle or 16 nM panobinostat for the indicated time. Percentage of cells in S phase (EdU positive) was determined by flow cytometry. Mean values of 2 independent experiments are shown; error bars represent standard deviation (SD). (B) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with vehicle (D) or 16 nM panobinostat (P) for the indicated time using antibodies to p16INK4A, p21WAF1/CIP1, and phosphorylated RB. β-actin served as loading control. The results shown are representative of 3 independent experiments. (C) Flow cytometry analysis of c-Kit expression in A/E9a;NrasG12D leukemic cells treated in vitro with 16 nM panobinostat for the indicated time. Mean values of 3 independent experiments are shown; error bars represent SD. (D) Quantitative real-time PCR of relative mRNA levels of target genes in A/E9a;NrasG12D leukemic cells treated in vitro with 16 nM panobinostat for 24 hours. Results were normalized to HPRT mRNA. Mean value of 3 to 6 individual experiments is shown, and error bars represent SD. (E) Quantitative real-time PCR of relative mRNA levels of target genes in GFP-positive splenocytes isolated from A/E9a;NrasG12D leukemia recipient mice 72 hours after initiation of treatment with panobinostat (25 mg/kg) or vehicle (D5W). Results were normalized to HPRT mRNA. Mean value of 3 to 5 individual samples is shown, and error bars represent standard error of the mean. (F-G) Flow cytometry analysis of c-Kit (F) and Mac-1 (G) expression in GFP-positive bone marrow cells isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W) for 3 days. Data are combined from 2 individual experiments. Each data point represents an individual mouse, and horizontal bars represent mean value. (F) *P = .0248; ***P = .0009. (G) *P = .0292. (H) Light microscopy of May-Grunwald/Giemsa–stained bone marrow cells isolated from A/E9a;NrasG12D leukemia recipient mice cells treated with panobinostat (25 mg/kg) or vehicle (D5W, 250 µL) for 5 days. Imaging was performed with a ×60 objective. Representative images of 5 biological replicates are shown (bars represent 10 µm). GFP-positive cells isolated from 5-day vehicle-treated mice (left panel) demonstrate immature blast morphology, including a fine rim of agranular basophilic cytoplasm with a round to oval nucleus containing “open chromatin” and 1 or more prominent nucleoli (arrowheads). In contrast, GFP-positive cells from 5-day panobinostat-treated mice (right panel) show features of maturation including a reduction in the nuclear:cytoplasmic ratio, condensation of nuclear chromatin, and infrequent nucleolation. Frequent coarse azurophilic cytoplasmic granules (arrowheads) indicate myeloid differentiation.
Panobinostat treatment of A/E9a;NrasG12Dleukemic cells triggers differentiation. (A) Cell cycle analysis of A/E9a;NrasG12D leukemic cells treated in vitro with vehicle or 16 nM panobinostat for the indicated time. Percentage of cells in S phase (EdU positive) was determined by flow cytometry. Mean values of 2 independent experiments are shown; error bars represent standard deviation (SD). (B) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with vehicle (D) or 16 nM panobinostat (P) for the indicated time using antibodies to p16INK4A, p21WAF1/CIP1, and phosphorylated RB. β-actin served as loading control. The results shown are representative of 3 independent experiments. (C) Flow cytometry analysis of c-Kit expression in A/E9a;NrasG12D leukemic cells treated in vitro with 16 nM panobinostat for the indicated time. Mean values of 3 independent experiments are shown; error bars represent SD. (D) Quantitative real-time PCR of relative mRNA levels of target genes in A/E9a;NrasG12D leukemic cells treated in vitro with 16 nM panobinostat for 24 hours. Results were normalized to HPRT mRNA. Mean value of 3 to 6 individual experiments is shown, and error bars represent SD. (E) Quantitative real-time PCR of relative mRNA levels of target genes in GFP-positive splenocytes isolated from A/E9a;NrasG12D leukemia recipient mice 72 hours after initiation of treatment with panobinostat (25 mg/kg) or vehicle (D5W). Results were normalized to HPRT mRNA. Mean value of 3 to 5 individual samples is shown, and error bars represent standard error of the mean. (F-G) Flow cytometry analysis of c-Kit (F) and Mac-1 (G) expression in GFP-positive bone marrow cells isolated from A/E9a;NrasG12D leukemia recipient mice treated with panobinostat (25 mg/kg) or vehicle (D5W) for 3 days. Data are combined from 2 individual experiments. Each data point represents an individual mouse, and horizontal bars represent mean value. (F) *P = .0248; ***P = .0009. (G) *P = .0292. (H) Light microscopy of May-Grunwald/Giemsa–stained bone marrow cells isolated from A/E9a;NrasG12D leukemia recipient mice cells treated with panobinostat (25 mg/kg) or vehicle (D5W, 250 µL) for 5 days. Imaging was performed with a ×60 objective. Representative images of 5 biological replicates are shown (bars represent 10 µm). GFP-positive cells isolated from 5-day vehicle-treated mice (left panel) demonstrate immature blast morphology, including a fine rim of agranular basophilic cytoplasm with a round to oval nucleus containing “open chromatin” and 1 or more prominent nucleoli (arrowheads). In contrast, GFP-positive cells from 5-day panobinostat-treated mice (right panel) show features of maturation including a reduction in the nuclear:cytoplasmic ratio, condensation of nuclear chromatin, and infrequent nucleolation. Frequent coarse azurophilic cytoplasmic granules (arrowheads) indicate myeloid differentiation.
To confirm that panobinostat was inducing tumor cell differentiation, the effect of panobinostat on A/E9a;NrasG12D leukemias in vivo over a 7-day time course demonstrated increased levels of GATA-2, SCL, and C/EBPα in GFP-positive A/E9a;NrasG12D leukemic cells 72 hours after exposure to the compound (Figure 5E and supplemental Figure 8). Flow cytometry demonstrated a time-dependent decrease of GFP-positive A/E9a;NrasG12D leukemic cells expressing c-Kit (Figure 5F) concomitant with an increase in cells expressing the myeloid differentiation marker Mac-1 (Figure 5G). Histologic analysis revealed A/E9a;NrasG12D leukemic cells from panobinostat-treated leukemic mice displaying a more granulocyte/macrophage-like appearance (Figure 5H).
Panobinostat mediates proteasome-dependent degradation of A/E9a
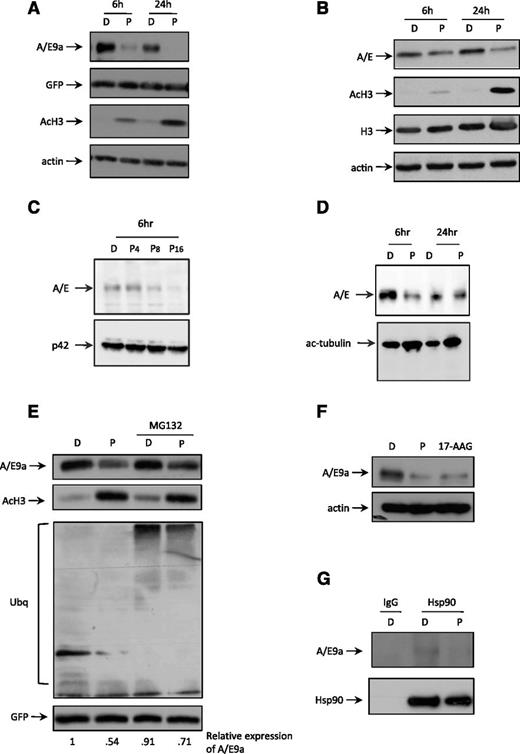
It has previously been observed that treatment of human Kasumi-1 cells with the class I–selective HDACi romidepsin resulted in apoptosis and reduced expression of A/E.33 Treatment of A/E9a;NrasG12D leukemic blasts with panobinostat in vitro for as little as 6 hours caused a marked decrease in expression of A/E9a concomitant with histone H3 hyperacetylation (Figure 6A). Importantly, there was no change in expression of GFP, which in our experimental system is coexpressed with A/E9a (Figure 6A), and there was no substantial change in the levels of A/E9a mRNA over a 24-hour time course (data not shown). We next treated Kasumi-1 cells with panobinostat and demonstrated a similar decrease in A/E with kinetics similar to those observed using the mouse A/E9a;NrasG12D leukemias (Figure 6B). In addition, primary human t(8;21) AML cells treated with panobinostat also showed time-dependent decrease in A/E (Figure 6C-D). These data demonstrate that panobinostat induces degradation of full-length A/E and the A/E9a fusion protein with similar kinetics. Cotreatment of A/E9a;NrasG12D leukemic cells with panobinostat and the proteasome inhibitor MG132 partially rescued the panobinostat-mediated decrease in A/E9a (Figure 6E), suggesting that panobinostat triggered proteasome-mediated decay of A/E9a. A/E has previously been shown to be a client protein for the molecular chaperone Hsp90,33 suggesting that HDACi-mediated hyperacetylation of Hsp90 may be sufficient to mediate degradation of A/E9a. Treatment of A/E9a;NrasG12D leukemic blasts in vitro with the Hsp90 inhibitor 17-N-Allylamino-17-demethoxygeldanamycin (17-AAG) resulted in a rapid loss in expression of A/E9a (Figure 6F), supporting previous studies demonstrating that that A/E is a client protein of Hsp9033 and that disruption in the association of A/E9a with Hsp90 may trigger its degradation. Immunoprecipitation assays showed that A/E9a physically interacted with Hsp90, which was diminished upon treatment with panobinostat (Figure 6G) and marginally reduced following treatment with 17-AAG (supplemental Figure 9). Taken together, these data demonstrate that A/E9a is an Hsp90 client protein and that treatment with panobinostat promotes dissociation of A/E9a from Hsp90 and supports a model in which hyperacetylation of Hsp90 results in subsequent ubiquitination and proteasomal degradation of A/E9a.
Panobinostat induces degradation of A/E9a and A/E. (A) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with DMSO vehicle (D) or 16 nM panobinostat (P) for the indicated time using antibodies to AML1, GFP, and acetylated histone H3. β-actin served as loading control. The results shown are representative of at least 3 independent experiments. (B) Western blot analysis of whole-cell lysates prepared from Kasumi cells treated in vitro with DMSO vehicle (D) or 8 nM panobinostat (P) for the indicated time using antibodies to AML1 and acetylated histone H3. Total histone H3 and β-actin served as loading controls. The results shown are representative of at least 3 independent experiments. (C) Western blot analysis of whole-cell lysates prepared from primary t(8;21) AML cells treated in vitro with DMSO (D) or indicated concentrations (nM) of panobinostat (P) for 6 hours using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for p42 as loading control (bottom panel). (D) Western blot analysis of whole-cell lysates prepared from a different primary t(8;21) AML sample as that shown in panel C treated in vitro with DMSO (D) or 8 nM panobinostat (P) for the indicated time using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for anti–acetyl-tubulin antibody (bottom panel). (E) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with DMSO vehicle (D) or 16 nM panobinostat (P) for 24 hours with addition of 5 µM MG132 for the final 4 hours (lanes 1 and 2) using antibodies to AML1, acetylated histone H3, ubiquitin, and GFP. The results shown are representative of at least 3 independent experiments. (F) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with vehicle (D), 16 nM panobinostat (P), or 100 nM of the Hsp90 inhibitor 17-AAG for 24 hours using antibodies to AML1. β-actin served as loading control. The results shown are representative of at least 3 independent experiments. (G) Western blot analysis of the interaction between A/E9a and Hsp90 in A/E9a;NrasG12D leukemic cells treated with vehicle (D) or 16 nM panobinostat (P) for 4 hours. A control mouse immunoglobulin G (IgG) and antibody to Hsp90 were used for immunoprecipitation; antibodies to AML1 and Hsp90 were used for western blotting. The results shown are representative of at least 3 independent experiments.
Panobinostat induces degradation of A/E9a and A/E. (A) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with DMSO vehicle (D) or 16 nM panobinostat (P) for the indicated time using antibodies to AML1, GFP, and acetylated histone H3. β-actin served as loading control. The results shown are representative of at least 3 independent experiments. (B) Western blot analysis of whole-cell lysates prepared from Kasumi cells treated in vitro with DMSO vehicle (D) or 8 nM panobinostat (P) for the indicated time using antibodies to AML1 and acetylated histone H3. Total histone H3 and β-actin served as loading controls. The results shown are representative of at least 3 independent experiments. (C) Western blot analysis of whole-cell lysates prepared from primary t(8;21) AML cells treated in vitro with DMSO (D) or indicated concentrations (nM) of panobinostat (P) for 6 hours using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for p42 as loading control (bottom panel). (D) Western blot analysis of whole-cell lysates prepared from a different primary t(8;21) AML sample as that shown in panel C treated in vitro with DMSO (D) or 8 nM panobinostat (P) for the indicated time using an antibody to AML1 (upper panel). Membrane was stripped and reprobed for anti–acetyl-tubulin antibody (bottom panel). (E) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with DMSO vehicle (D) or 16 nM panobinostat (P) for 24 hours with addition of 5 µM MG132 for the final 4 hours (lanes 1 and 2) using antibodies to AML1, acetylated histone H3, ubiquitin, and GFP. The results shown are representative of at least 3 independent experiments. (F) Western blot analysis of whole-cell lysates prepared from A/E9a;NrasG12D leukemic cells treated in vitro with vehicle (D), 16 nM panobinostat (P), or 100 nM of the Hsp90 inhibitor 17-AAG for 24 hours using antibodies to AML1. β-actin served as loading control. The results shown are representative of at least 3 independent experiments. (G) Western blot analysis of the interaction between A/E9a and Hsp90 in A/E9a;NrasG12D leukemic cells treated with vehicle (D) or 16 nM panobinostat (P) for 4 hours. A control mouse immunoglobulin G (IgG) and antibody to Hsp90 were used for immunoprecipitation; antibodies to AML1 and Hsp90 were used for western blotting. The results shown are representative of at least 3 independent experiments.
Inducible deletion of A/E9a phenocopies the effect of panobinostat
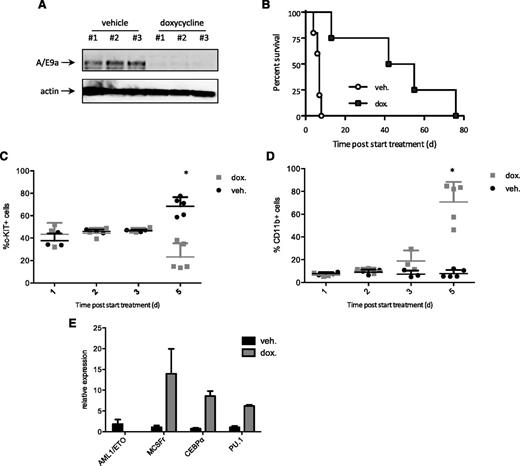
To determine if genetic deletion of A/E9a phenocopied the effects of panobinostat, we established a model of t(8;21) driven by doxycycline-inducible expression of A/E9a and constitutive expression of oncogenic Nras. Mice were transplanted with Nzeg-eGFP fetal liver cells transduced with 2 retroviral vectors, 1 encoding dsRed-linked A/E9a under the control of a tetracycline response element (TREtight) promoter, and the other encoding NrasG12D coexpressing the ‘‘Tet-off’’ tetracycline transactivator, which shuts off the expression of TRE-regulated genes in a doxycycline-dependent manner. Primary GFP-positive, dsRed-positive Tet-off A/E9a;NrasG12D leukemias were established in mice, which were then treated with doxycycline or vehicle. In vivo exposure to doxycycline efficiently silenced A/E9a and dsRed protein (Figure 7A and supplemental Figure 10A-C), leading to a substantial survival advantage compared with vehicle-treated mice (Figure 7B). Mice that relapse while being treated with doxycycline present with morphologic features of A/E9a;NrasG12D leukemia including the presence of GFP-positive, dsRed-positive cells in the peripheral blood and bone marrow (Ghisi and Johnstone, unpublished observations). This suggests that the Tet-off system used to express A/E9a in a regulated manner is not 100% effective in a small proportion of cells, and these cells eventually grow out, resulting in the death of the mouse.
Inducible deletion of A/E9a phenocopies the effect of panobinostat. (A) Western blot analysis of whole-cell lysates prepared from GFP-positive spleen cells isolated from Tet-off A/E9a;NrasG12D leukemia recipient mice 72 hours after initiation of treatment with doxycycline or vehicle using an antibody to AML1. β-actin served as loading control. The results shown are representative of 2 independent experiments. (B) Kaplan-Meier survival curves of treated Tet-off A/E9a;NrasG12D leukemia recipient mice following initiation of therapy (n = 5 for vehicle, n = 4 for doxycycline; median survival benefit 42 days, P = .0051). (C-D) Flow cytometry analysis of c-Kit and CD11b expression in donor GFP-positive spleen cells isolated from Tet-off A/E9a;NrasG12D leukemia recipient mice at the indicated time after initiation of treatment with doxycycline (2 mg/kg) or vehicle. Each data point represents an individual mouse, and the mean value ± standard error from 2 separate experiments is shown. *P < .001. (E) Mice bearing Tet-off A/E9a;NrasG12D leukemias were treated with vehicle or doxycycline (dox) for 5 days. Tumors were harvested, and expression of the indicated genes was determined by quantitative real-time PCR. Results (mean and standard error of the mean) shown are from tumors harvested from 5 individual recipient mice for each treatment regimen.
Inducible deletion of A/E9a phenocopies the effect of panobinostat. (A) Western blot analysis of whole-cell lysates prepared from GFP-positive spleen cells isolated from Tet-off A/E9a;NrasG12D leukemia recipient mice 72 hours after initiation of treatment with doxycycline or vehicle using an antibody to AML1. β-actin served as loading control. The results shown are representative of 2 independent experiments. (B) Kaplan-Meier survival curves of treated Tet-off A/E9a;NrasG12D leukemia recipient mice following initiation of therapy (n = 5 for vehicle, n = 4 for doxycycline; median survival benefit 42 days, P = .0051). (C-D) Flow cytometry analysis of c-Kit and CD11b expression in donor GFP-positive spleen cells isolated from Tet-off A/E9a;NrasG12D leukemia recipient mice at the indicated time after initiation of treatment with doxycycline (2 mg/kg) or vehicle. Each data point represents an individual mouse, and the mean value ± standard error from 2 separate experiments is shown. *P < .001. (E) Mice bearing Tet-off A/E9a;NrasG12D leukemias were treated with vehicle or doxycycline (dox) for 5 days. Tumors were harvested, and expression of the indicated genes was determined by quantitative real-time PCR. Results (mean and standard error of the mean) shown are from tumors harvested from 5 individual recipient mice for each treatment regimen.
As seen following in vivo treatment of mice bearing A/E9a;NrasG12D leukemias with panobinostat, genetic depletion of A/E9a was accompanied by a decrease in c-Kit–positive cells (Figure 7C), an increase in Mac-1–positive cells (Figure 7D), and an increase in genes (M/CSF receptor, C/EBPα and PU.1) associated with myeloid differentiation (Figure 7E). Moreover, histologic analysis revealed that A/E9a;NrasG12D leukemic cells harvested from doxycycline-treated leukemic mice assumed a differentiated granulocyte/macrophage-like morphology similar to that observed following treatment with panobinostat (supplemental Figure 10D).These data demonstrate that a reduction in the expression of A/E9a in primary leukemias phenocopies the molecular, biological, and therapeutic effects of panobinostat treatment.
Discussion
Aberrant recruitment of HDACs by A/E provides a strong molecular rational to use HDACis to treat t(8;21) AML. Initial studies aimed to address this hypothesis predominantly used in vitro assays, a single human cell line (Kasumi), and a weak HDACi (VPA) that needs to be used at millimolar concentrations to induce histone hyperacetylation.30,32 Although these studies hinted that an HDACi may be effective in this setting, therapeutic insights were lacking, and it was unclear if tumor cell apoptosis or differentiation was the primary effector mechanism of HDACis.32,33 Herein, we describe the therapeutic, biological, and molecular effects of the HDACis vorinostat and panobinostat in an established and transplantable mouse model of t(8;21) AML. Panobinostat was effective in reducing tumor burden in A/E9a;NrasG12D leukemia–bearing mice, whereas those transplanted with leukemias expressing MLL/ENL;NrasG12D or MLL/AF9;NrasG12D were more resistant to panobinostat therapy. Interestingly, the primary biological response to panobinostat in A/E9a;NrasG12D leukemia was induction of differentiation rather than apoptosis, preceded by degradation of the A/E9a fusion protein through the release of A/E9a from Hsp90. Furthermore, we used an inducible A/E9a expression system to demonstrate that established A/E9a;NrasG12D leukemias were dependent on A/E9a for their long-term survival, and that genetic depletion of A/E9a phenocopied the biological and therapeutic effects of panobinostat. These results provide important insight into the working mechanism of HDACis in A/E9a;NrasG12D-driven AML, highlight the continued dependence of A/E9a;NrasG12D leukemias on the presence of A/E9a, and imply that targeting of this oncogenic fusion protein and using its expression as a biomarker for response to HDACis may have important implications for future clinical development.
Acute promyelocytic leukemia driven by the PML/RARα fusion protein is a primary example in which the relationship between clinical outcome and therapy-induced degradation of a single oncoprotein is evident.48,49 Although treatment of t(15;17) acute promyelocytic leukemia with retinoic acid (RA) results in differentiation and leukemia regression, this monotherapy rarely leads to prolonged remissions. Combination with arsenic, however, clearly improves clinical efficacy of RA, and the key determinant to this clinical success may be the loss of leukemia-initiating cells through the degradation of PML/RARA.50,51 Similar improvements in therapeutic responses have also been observed for the combination of RA with VPA, which correlated with more pronounced degradation of PML/RARA.52 Our data show that in A/E9a;NrasG12D-driven leukemia, treatment with panobinostat as monotherapy results in a therapeutic response that is associated with myeloid differentiation and degradation of A/E9a. We noted that upon cessation of panobinostat treatment, mice relapsed with disease that was pathologically and phenotypically similar to the original disease. We posit that using the regime undertaken in this study, panobinostat was unable to induce differentiation of all A/E9a;NrasG12D tumors, and mice therefore subsequently relapsed. This is supported by our studies demonstrating that panobinostat remained fully effective in mice transplanted with A/E9a;NrasG12D cells harvested from mice previously treated with panobinostat (supplemental Figure 1E). Moreover, FACS analysis of A/E9a;NrasG12D cells harvested from mice treated with vehicle or panobinostat for 4 weeks revealed no change in hemopoietic cell surface markers (supplemental Figure 1D). Finally, we have performed studies showing that transplantation of only 1 A/E9a;NrasG12D tumor into recipient mice was sufficient to induce fatal disease in recipient mice.
Depletion of A/E9a in established leukemias phenocopied the effect of panobinostat, supporting a model in which HDACi-mediated degradation of A/E9a is the primary biological event involved in the elimination of leukemic cells and the therapeutic response observed. As expression of other oncogenes, such as NrasG12D, was maintained in the experimental systems used, this implies that A/E9a;NrasG12D AMLs are addicted to A/E9a and that other compounds such as Hsp90 antagonists that indirectly target A/E9a may also be therapeutically effective. Interestingly and in contrast to A/E9a, MLL fusion proteins do not require Hsp90 as a chaperone, and as such, expression levels of MLL fusion proteins are not affected upon treatment with 17-AAG.53 Given the mechanism of action of panobinostat in A/E9a;NrasG12D AMLs, this may explain why the therapeutic responses observed in our MLL fusion protein–expressing AMLs were relatively poor.
Previous studies proposed that tumor-specific induction of the TRAIL death receptor pathway was responsible for the antitumor effects of HDACis.31,46 Consistent with those studies, we found that Dr5 was transcriptionally activated following treatment of A/E9a;NrasG12D leukemias with panobinostat. However, A/E9a;NrasG12D leukemias with genetic knockout of either Dr5 or Trail were equally sensitive to panobinostat treatment as unmodified A/E9a;NrasG12D leukemias, providing definitive genetic evidence that the TRAIL signaling pathway plays no role in mediating the antitumor response to HDACis in A/E9a-driven leukemia. In contrast to our previous studies using the Eμ-myc model of B-cell lymphoma, which demonstrated a direct link between activation of the intrinsic apoptosis pathway and therapeutic efficacy,43,44 we saw no evidence of apoptosis following treatment of A/E9a tumors with panobinostat. These findings indicate that the molecular and biological effector mechanisms of HDACis identified for one tumor type may not be directly translatable to another and highlight the importance of testing anticancer drugs in physiologically relevant and genetically tractable preclinical models of disease. Panobinostat treatment also provided a significant survival benefit to mice bearing A/E9a;NrasG12D/p53−/− leukemias; however, the survival benefit was less pronounced than that observed using A/E9a;NrasG12D with WT p53. We and others have demonstrated that a functional p53 is not required for HDACis to induce tumor cell apoptosis (see Bolden et al10 and references therein). However, it is possible that p53 is required for HDACis to induce a robust differentiation response as has been recently demonstrated for the treatment of MLL-ENL/HrasV12D–driven AML treated with DNA-damaging agents.54
Although A/E has been shown to be a direct target of HATs and in fact the transforming activity of A/E is dependent on p300-mediated acetylation of the fusion protein,27 we saw no change in the acetylation status of A/E9a following HDACi treatment (data not shown). Our finding that HDACi-induced degradation of A/E9a in primary leukemic cells is mediated through release of A/E9a from the molecular chaperone Hsp90 is consistent with previous observations using HDACi in the human t(8;21) cell line Kasumi-1.33 It has been proposed that the chaperone activity of Hsp90 is regulated through deacetylation by HDAC655 and that HDACi-induced hyperacetylation of Hsp90 then leads to release and subsequent proteasome-mediated degradation of client proteins such as A/E9a. However, it is controversial whether HDAC6-specific deacetylation of Hsp90 plays an important role in stabilizing the A/E9a protein. Using romidepsin, a very weak inhibitor of HDAC6, we still observed degradation of A/E9a (data not shown). This suggests that inhibition of class I HDACs is sufficient to mediate degradation of A/E9a and is consistent with recent studies by us and others demonstrating that class I HDACs can regulate the acetylation status of Hsp90.56
In summary, we demonstrated that the therapeutic efficiency of HDACis in A/E9a AML is mediated by degradation of A/E9a and subsequent differentiation of leukemic cells along the myeloid lineage. Our data showing therapeutic responses in leukemias with knockout of p53, Dr5, or Trail, or overexpressing the antiapoptotic protein Bcl-2, provide preclinical evidence that panobinostat will be effective for the treatment of t(8;21) AML and will have a clear advantage over conventional therapeutic agents in leukemias that have acquired resistance toward apoptosis, such as leukemias deficient for p53. Our findings, further supported by the positive responses previously observed in t(8;21) patients treated with HDACi,14 indicate that HDACi-mediated differentiation therapy is an attractive and molecularly rational treatment strategy for this type of cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Leonie Cluse, Edwin Hawkins, and Geoff Matthews for constructive criticism and helpful advice, and Klaus Matthaei for the Nzeg-eGFP mice.
This work was supported by fellowships from the Dutch Cancer Society (AMC2009-4457) (M.B.) and (NKI2009-4446) (I.V.). R.W.J. is a Principal Research Fellow of the National Health and Medical Research Council of Australia (NHMRC) and is supported by NHMRC Program and Project Grants, the Susan G. Komen Breast Cancer Foundation, Cancer Council Victoria, the Victorian Cancer Agency, and the Leukemia Foundation of Australia.
Authorship
Contribution: M.B. designed research, performed research, collected, analyzed, and interpreted data, performed statistical analysis, and wrote the manuscript; I.V., B.P.M., J.M.S., M.G., K.S., A.B., and J.S. performed research and collected, analyzed, and interpreted data; J.Z. designed research and contributed vital new reagents; A.R.R., P.A., G.J.O., and S.W.L. contributed vital new reagents; R.W.J. designed research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: P.A. is a paid employee of Novartis Pharmaceuticals, which provides grant support to the laboratory of R.W.J. The remaining authors declare no competing financial interests.
The current affiliation for M.B. is Laboratory for Experimental Oncology and Radiobiology, Center for Experimental Molecular Medicine, Academic Medical Center, Amsterdam, The Netherlands.
The current affiliation for I.V. is Division of Immunology, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
The current affiliation for J.Z. is Research Institute of Molecular Pathology, Dr Bohr-Gasse 7, A-1030 Vienna, Austria.
Correspondence: Ricky W. Johnstone, Cancer Therapeutics Program, Peter MacCallum Cancer Centre, St. Andrews Place, East Melbourne 3002, Victoria, Australia; e-mail: ricky.johnstone@petermac.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal